Gene
KWMTBOMO10040 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005528
Annotation
methylglutaconyl-CoA_hydratase_[Agrotis_segetum]
Full name
Enoyl-CoA hydratase domain-containing protein 2, mitochondrial
Location in the cell
Mitochondrial Reliability : 2.018
Sequence
CDS
ATGTTATTAAGTAAATTAAAATTGAGATCCTTCATTGTTAGAGTTGTAAATACCCGAAATCTAGCCACAAAGATACAACAACTGAACGAAAATGTGAATCCTGTTGTATTTGAGAAACTTACTGGGGTAGATAAGGGTATCGCATTATGCGGTCTCAACAGCCCTAAAGACAGGAACGCACTCGGATTTACCCTCATAGATGCGATGCGCGAGGTCAACCAGATCATTCGAGAAGATACCAAACTGTCTGTTGTTATCTTTCATAGTATGGTTCCGGGAATTTTCTGCGCTGGAGCTAACCTAAAAGAACGCTTAAAAATGTCCGATGAAGAGGTAGCCAAATTCGTTCGAGGGCTTAGAGAAACTTTTATTGAAATCGAAGACTTGCCCATGCCAACCATAGCTGCTGTGGAAGGAGTGGCTGTTGGTGGAGGGTTGGAATTAGCCTTGGCATGTGACATCAGGATAGCAGCAGATACTGCTAAACTAGGTCTTGTCGAGACTGGTAGAGGTCTTATTCCCGGTGCTGGAGGTACCCAGAGACTACCAAGAACTATTCAGCTACCAATTGCTAAAGAGTTGATATTTACATCAAGAATTGTTAGTGGAAAGGAAGCTAAAGCATTAGGCATAGTGAATCATGTGGTCGCACAAGACACTGCAAACAAAGCAGCCTTTGAGAAGGCCTTGAGCATTGCACGAGAAATCATCCCGAATGCCCCCATAGCCCTCCGTTGTGCAAAACAAGCTATCAGTGAAGGCATACAGCTGAGTATCAAGGATGGTTATGAGATAGAACAGAAATGCTACGAAATAAATATTCCAACTAAAGACAGACAGGAAGGCATGATTTCATTTATGGAAAAAAGAAAACCTGTTTATGAGGGACATTAG
Protein
MLLSKLKLRSFIVRVVNTRNLATKIQQLNENVNPVVFEKLTGVDKGIALCGLNSPKDRNALGFTLIDAMREVNQIIREDTKLSVVIFHSMVPGIFCAGANLKERLKMSDEEVAKFVRGLRETFIEIEDLPMPTIAAVEGVAVGGGLELALACDIRIAADTAKLGLVETGRGLIPGAGGTQRLPRTIQLPIAKELIFTSRIVSGKEAKALGIVNHVVAQDTANKAAFEKALSIAREIIPNAPIALRCAKQAISEGIQLSIKDGYEIEQKCYEINIPTKDRQEGMISFMEKRKPVYEGH
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Fatty acid metabolism
Lipid metabolism
Lyase
Mitochondrion
Reference proteome
Transit peptide
Feature
chain Enoyl-CoA hydratase domain-containing protein 2, mitochondrial
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9J7N6
A0A2W1BDU2
A0A2H1WVB4
A0A2A4JK17
A0A068FRG3
A0A0N1IG39
+ More
A0A194QC21 A0A212FF89 A0A0L7LS28 A0A3B0JXX1 A0A1A9WQK7 Q293E8 A0A1A9V7K5 A0A1I8QAE9 D3TMQ1 A0A1B0BED9 A0A1I8MGD0 A0A1A9Y995 A0A0A1WDA8 A0A0K8TMJ8 A0A0K8VQX0 A0A034VHU1 W8BT04 B4LP83 A0A1W4VPT6 B4KSP9 A0A182MUN3 B4MRY7 A1Z934 B4P506 B3MCU5 B4QDE9 A0A154P623 A0A088A241 A0A2A3E121 V9IJN7 B4JVM4 A0A151K0K2 B3NRX2 F4W715 A0A0B4J2T1 A0A232F961 A0A0N0U4J9 A0A195C3R8 A0A195AXN7 A0A158NM99 Q6DC25 A0A2D0SRC8 F1RDX1 A0A151X822 A0A0M4EWD6 A0A182QA34 A0A182KB30 A0A182FBY1 A0A0L7R4P2 A0A182JN03 A0A182LH73 A0A1S4H6F3 W5JDL7 A0A182TSI0 A0A2M4BV91 A0A2M4BUV0 A0A093QF19 A0A084VHY8 A0A3B4CQ36 A0A182Y722 A0A182V9B6 A0A182SH58 A0A3B3QCT0 R4WJY7 A0A218VEC6 A0A2C9GRT1 A0A2D0Q454 A0A0L0CS03 A0A3B5APY0 M4ANX5 A0A099Z015 A0A2M4AU08 A0A2M3ZAK0 A0A0B6Y9E6 A0A3P9NA04 A0A151M4H3 A0A1V4JY92 W5MNK4 A0A3P8YLY9 Q3TLP5 H2THP1 A0A3Q1EFY0 A0A0J7KTU4 A0A3L8SPZ4 A0A3Q3AI09 F1NSS6 A0A2R7WRU6 A0A2I4BS85 A0A3Q3J982 A0A182S090 A0A3Q1I191 A0A026WGH0 A0A3Q1G057 A0A146QKP3
A0A194QC21 A0A212FF89 A0A0L7LS28 A0A3B0JXX1 A0A1A9WQK7 Q293E8 A0A1A9V7K5 A0A1I8QAE9 D3TMQ1 A0A1B0BED9 A0A1I8MGD0 A0A1A9Y995 A0A0A1WDA8 A0A0K8TMJ8 A0A0K8VQX0 A0A034VHU1 W8BT04 B4LP83 A0A1W4VPT6 B4KSP9 A0A182MUN3 B4MRY7 A1Z934 B4P506 B3MCU5 B4QDE9 A0A154P623 A0A088A241 A0A2A3E121 V9IJN7 B4JVM4 A0A151K0K2 B3NRX2 F4W715 A0A0B4J2T1 A0A232F961 A0A0N0U4J9 A0A195C3R8 A0A195AXN7 A0A158NM99 Q6DC25 A0A2D0SRC8 F1RDX1 A0A151X822 A0A0M4EWD6 A0A182QA34 A0A182KB30 A0A182FBY1 A0A0L7R4P2 A0A182JN03 A0A182LH73 A0A1S4H6F3 W5JDL7 A0A182TSI0 A0A2M4BV91 A0A2M4BUV0 A0A093QF19 A0A084VHY8 A0A3B4CQ36 A0A182Y722 A0A182V9B6 A0A182SH58 A0A3B3QCT0 R4WJY7 A0A218VEC6 A0A2C9GRT1 A0A2D0Q454 A0A0L0CS03 A0A3B5APY0 M4ANX5 A0A099Z015 A0A2M4AU08 A0A2M3ZAK0 A0A0B6Y9E6 A0A3P9NA04 A0A151M4H3 A0A1V4JY92 W5MNK4 A0A3P8YLY9 Q3TLP5 H2THP1 A0A3Q1EFY0 A0A0J7KTU4 A0A3L8SPZ4 A0A3Q3AI09 F1NSS6 A0A2R7WRU6 A0A2I4BS85 A0A3Q3J982 A0A182S090 A0A3Q1I191 A0A026WGH0 A0A3Q1G057 A0A146QKP3
Pubmed
19121390
28756777
26385554
26354079
22118469
26227816
+ More
15632085 17994087 20353571 25315136 25830018 26369729 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 21719571 20075255 28648823 21347285 23594743 20966253 12364791 20920257 23761445 24438588 25244985 29240929 23691247 26108605 23542700 22293439 25069045 16141072 19468303 15489334 21183079 23806337 23576753 21551351 30282656 15592404 24508170 30249741
15632085 17994087 20353571 25315136 25830018 26369729 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 21719571 20075255 28648823 21347285 23594743 20966253 12364791 20920257 23761445 24438588 25244985 29240929 23691247 26108605 23542700 22293439 25069045 16141072 19468303 15489334 21183079 23806337 23576753 21551351 30282656 15592404 24508170 30249741
EMBL
BABH01020426
KZ150179
PZC72531.1
ODYU01011325
SOQ57003.1
NWSH01001312
+ More
PCG71742.1 KJ622095 AID66685.1 KQ460044 KPJ18299.1 KQ459232 KPJ02535.1 AGBW02008849 OWR52390.1 JTDY01000229 KOB78184.1 OUUW01000001 SPP75938.1 CM000071 EAL24663.2 CCAG010005873 EZ422703 ADD18979.1 JXJN01012868 GBXI01017450 JAC96841.1 GDAI01002227 JAI15376.1 GDHF01011033 JAI41281.1 GAKP01016081 JAC42871.1 GAMC01002100 JAC04456.1 CH940648 EDW60192.2 CH933808 EDW10548.1 AXCM01001158 CH963850 EDW74876.1 AE013599 BT031029 AAF58477.2 ABV82411.1 CM000158 EDW90727.1 CH902619 EDV37347.1 CM000362 CM002911 EDX06814.1 KMY93296.1 KQ434823 KZC07307.1 KZ288467 PBC25415.1 JR049120 AEY60867.1 CH916375 EDV98492.1 KQ981253 KYN44174.1 CH954179 EDV56274.1 GL887802 EGI70005.1 AAZX01007340 NNAY01000614 OXU27391.1 KQ435812 KOX72771.1 KQ978344 KYM94823.1 KQ976716 KYM76807.1 ADTU01002232 BC078266 AAH78266.1 CR848004 KQ982431 KYQ56474.1 CP012524 ALC42095.1 AXCN02001118 KQ414657 KOC65835.1 AAAB01008816 ADMH02001689 ETN61423.1 GGFJ01007710 MBW56851.1 GGFJ01007709 MBW56850.1 KL671494 KFW82677.1 ATLV01013250 KE524847 KFB37582.1 AK417930 BAN21145.1 MUZQ01000002 OWK64444.1 APCN01004488 JRES01000091 KNC34204.1 KL886654 KGL74125.1 GGFK01010936 MBW44257.1 GGFM01004749 MBW25500.1 HACG01005210 CEK52075.1 AKHW03006631 KYO19371.1 LSYS01005497 OPJ77075.1 AHAT01002719 AHAT01002720 AHAT01002721 AHAT01002722 AHAT01002723 AK005030 AK153878 AK160482 AK166388 AK166965 AL844206 BX293563 BC025104 LBMM01003274 KMQ93721.1 QUSF01000010 RLW05969.1 AADN05000647 KK855346 PTY22091.1 KK107231 QOIP01000009 EZA55068.1 RLU18948.1 GCES01129614 GCES01055082 JAQ56708.1 JAR31241.1
PCG71742.1 KJ622095 AID66685.1 KQ460044 KPJ18299.1 KQ459232 KPJ02535.1 AGBW02008849 OWR52390.1 JTDY01000229 KOB78184.1 OUUW01000001 SPP75938.1 CM000071 EAL24663.2 CCAG010005873 EZ422703 ADD18979.1 JXJN01012868 GBXI01017450 JAC96841.1 GDAI01002227 JAI15376.1 GDHF01011033 JAI41281.1 GAKP01016081 JAC42871.1 GAMC01002100 JAC04456.1 CH940648 EDW60192.2 CH933808 EDW10548.1 AXCM01001158 CH963850 EDW74876.1 AE013599 BT031029 AAF58477.2 ABV82411.1 CM000158 EDW90727.1 CH902619 EDV37347.1 CM000362 CM002911 EDX06814.1 KMY93296.1 KQ434823 KZC07307.1 KZ288467 PBC25415.1 JR049120 AEY60867.1 CH916375 EDV98492.1 KQ981253 KYN44174.1 CH954179 EDV56274.1 GL887802 EGI70005.1 AAZX01007340 NNAY01000614 OXU27391.1 KQ435812 KOX72771.1 KQ978344 KYM94823.1 KQ976716 KYM76807.1 ADTU01002232 BC078266 AAH78266.1 CR848004 KQ982431 KYQ56474.1 CP012524 ALC42095.1 AXCN02001118 KQ414657 KOC65835.1 AAAB01008816 ADMH02001689 ETN61423.1 GGFJ01007710 MBW56851.1 GGFJ01007709 MBW56850.1 KL671494 KFW82677.1 ATLV01013250 KE524847 KFB37582.1 AK417930 BAN21145.1 MUZQ01000002 OWK64444.1 APCN01004488 JRES01000091 KNC34204.1 KL886654 KGL74125.1 GGFK01010936 MBW44257.1 GGFM01004749 MBW25500.1 HACG01005210 CEK52075.1 AKHW03006631 KYO19371.1 LSYS01005497 OPJ77075.1 AHAT01002719 AHAT01002720 AHAT01002721 AHAT01002722 AHAT01002723 AK005030 AK153878 AK160482 AK166388 AK166965 AL844206 BX293563 BC025104 LBMM01003274 KMQ93721.1 QUSF01000010 RLW05969.1 AADN05000647 KK855346 PTY22091.1 KK107231 QOIP01000009 EZA55068.1 RLU18948.1 GCES01129614 GCES01055082 JAQ56708.1 JAR31241.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000268350 UP000091820 UP000001819 UP000078200 UP000095300 UP000092444 UP000092460 UP000095301 UP000092443 UP000008792 UP000192221 UP000009192 UP000075883 UP000007798 UP000000803 UP000002282 UP000007801 UP000000304 UP000076502 UP000005203 UP000242457 UP000001070 UP000078541 UP000008711 UP000007755 UP000002358 UP000215335 UP000053105 UP000078542 UP000078540 UP000005205 UP000221080 UP000000437 UP000075809 UP000092553 UP000075886 UP000075881 UP000069272 UP000053825 UP000075880 UP000075882 UP000000673 UP000075902 UP000053258 UP000030765 UP000261440 UP000076408 UP000075903 UP000075901 UP000261540 UP000197619 UP000075840 UP000037069 UP000261400 UP000002852 UP000053641 UP000242638 UP000050525 UP000190648 UP000018468 UP000265140 UP000000589 UP000005226 UP000257200 UP000036403 UP000276834 UP000264800 UP000000539 UP000192220 UP000261600 UP000075900 UP000265040 UP000053097 UP000279307
UP000268350 UP000091820 UP000001819 UP000078200 UP000095300 UP000092444 UP000092460 UP000095301 UP000092443 UP000008792 UP000192221 UP000009192 UP000075883 UP000007798 UP000000803 UP000002282 UP000007801 UP000000304 UP000076502 UP000005203 UP000242457 UP000001070 UP000078541 UP000008711 UP000007755 UP000002358 UP000215335 UP000053105 UP000078542 UP000078540 UP000005205 UP000221080 UP000000437 UP000075809 UP000092553 UP000075886 UP000075881 UP000069272 UP000053825 UP000075880 UP000075882 UP000000673 UP000075902 UP000053258 UP000030765 UP000261440 UP000076408 UP000075903 UP000075901 UP000261540 UP000197619 UP000075840 UP000037069 UP000261400 UP000002852 UP000053641 UP000242638 UP000050525 UP000190648 UP000018468 UP000265140 UP000000589 UP000005226 UP000257200 UP000036403 UP000276834 UP000264800 UP000000539 UP000192220 UP000261600 UP000075900 UP000265040 UP000053097 UP000279307
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J7N6
A0A2W1BDU2
A0A2H1WVB4
A0A2A4JK17
A0A068FRG3
A0A0N1IG39
+ More
A0A194QC21 A0A212FF89 A0A0L7LS28 A0A3B0JXX1 A0A1A9WQK7 Q293E8 A0A1A9V7K5 A0A1I8QAE9 D3TMQ1 A0A1B0BED9 A0A1I8MGD0 A0A1A9Y995 A0A0A1WDA8 A0A0K8TMJ8 A0A0K8VQX0 A0A034VHU1 W8BT04 B4LP83 A0A1W4VPT6 B4KSP9 A0A182MUN3 B4MRY7 A1Z934 B4P506 B3MCU5 B4QDE9 A0A154P623 A0A088A241 A0A2A3E121 V9IJN7 B4JVM4 A0A151K0K2 B3NRX2 F4W715 A0A0B4J2T1 A0A232F961 A0A0N0U4J9 A0A195C3R8 A0A195AXN7 A0A158NM99 Q6DC25 A0A2D0SRC8 F1RDX1 A0A151X822 A0A0M4EWD6 A0A182QA34 A0A182KB30 A0A182FBY1 A0A0L7R4P2 A0A182JN03 A0A182LH73 A0A1S4H6F3 W5JDL7 A0A182TSI0 A0A2M4BV91 A0A2M4BUV0 A0A093QF19 A0A084VHY8 A0A3B4CQ36 A0A182Y722 A0A182V9B6 A0A182SH58 A0A3B3QCT0 R4WJY7 A0A218VEC6 A0A2C9GRT1 A0A2D0Q454 A0A0L0CS03 A0A3B5APY0 M4ANX5 A0A099Z015 A0A2M4AU08 A0A2M3ZAK0 A0A0B6Y9E6 A0A3P9NA04 A0A151M4H3 A0A1V4JY92 W5MNK4 A0A3P8YLY9 Q3TLP5 H2THP1 A0A3Q1EFY0 A0A0J7KTU4 A0A3L8SPZ4 A0A3Q3AI09 F1NSS6 A0A2R7WRU6 A0A2I4BS85 A0A3Q3J982 A0A182S090 A0A3Q1I191 A0A026WGH0 A0A3Q1G057 A0A146QKP3
A0A194QC21 A0A212FF89 A0A0L7LS28 A0A3B0JXX1 A0A1A9WQK7 Q293E8 A0A1A9V7K5 A0A1I8QAE9 D3TMQ1 A0A1B0BED9 A0A1I8MGD0 A0A1A9Y995 A0A0A1WDA8 A0A0K8TMJ8 A0A0K8VQX0 A0A034VHU1 W8BT04 B4LP83 A0A1W4VPT6 B4KSP9 A0A182MUN3 B4MRY7 A1Z934 B4P506 B3MCU5 B4QDE9 A0A154P623 A0A088A241 A0A2A3E121 V9IJN7 B4JVM4 A0A151K0K2 B3NRX2 F4W715 A0A0B4J2T1 A0A232F961 A0A0N0U4J9 A0A195C3R8 A0A195AXN7 A0A158NM99 Q6DC25 A0A2D0SRC8 F1RDX1 A0A151X822 A0A0M4EWD6 A0A182QA34 A0A182KB30 A0A182FBY1 A0A0L7R4P2 A0A182JN03 A0A182LH73 A0A1S4H6F3 W5JDL7 A0A182TSI0 A0A2M4BV91 A0A2M4BUV0 A0A093QF19 A0A084VHY8 A0A3B4CQ36 A0A182Y722 A0A182V9B6 A0A182SH58 A0A3B3QCT0 R4WJY7 A0A218VEC6 A0A2C9GRT1 A0A2D0Q454 A0A0L0CS03 A0A3B5APY0 M4ANX5 A0A099Z015 A0A2M4AU08 A0A2M3ZAK0 A0A0B6Y9E6 A0A3P9NA04 A0A151M4H3 A0A1V4JY92 W5MNK4 A0A3P8YLY9 Q3TLP5 H2THP1 A0A3Q1EFY0 A0A0J7KTU4 A0A3L8SPZ4 A0A3Q3AI09 F1NSS6 A0A2R7WRU6 A0A2I4BS85 A0A3Q3J982 A0A182S090 A0A3Q1I191 A0A026WGH0 A0A3Q1G057 A0A146QKP3
PDB
2ZQR
E-value=1.05619e-61,
Score=599
Ontologies
PATHWAY
GO
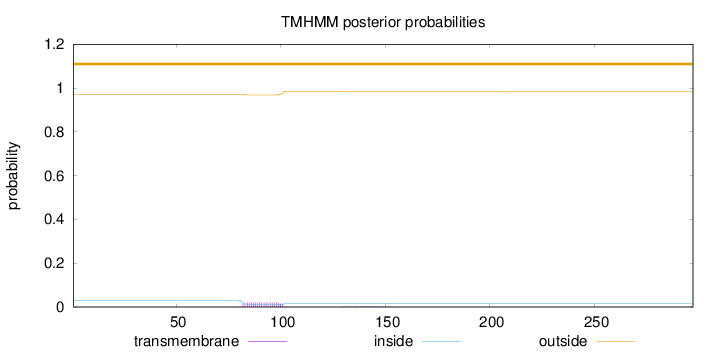
Topology
Subcellular location
Mitochondrion
Length:
297
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44488
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02872
outside
1 - 297
Population Genetic Test Statistics
Pi
167.003305
Theta
142.393223
Tajima's D
0.504978
CLR
83.465946
CSRT
0.517274136293185
Interpretation
Uncertain
