Gene
KWMTBOMO10039
Pre Gene Modal
BGIBMGA005527
Annotation
regulatory-associated_protein_of_TOR_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.176
Sequence
CDS
ATGACAGAAATGCCTCCTTATCACGCTGTTAAAGATGAAGAGAAAACCAGTGGTCAGAGTGACAGTGATTCCTCTAGCGATACTGATGATTGGGATTTATCTCAATGTTTTGATAAGCCGCAGCACCTTGAGCCTATAAAAGGCGCAGAACGGGTAGAGCACTCTTGGAGAGTTAAAGAAAAGTACAAGACACATTGCGTGGCCCTAGTTCTATGCCTGAATGTTGGCGTGGACCCTCCTGATGTTGTTAAAACACAACCCTGTGCGAGGCTCGAGTGTTGGATTGATCCTTTATCTTTATCACCGAGCAAGGCATTAGAAAGCATTGGTCATGCATTACAATCTCAGTATGAGAGATGGCAACCACGAGCCAGATACAAGCAATCTCTTGACCCCACTAGTGAAGAGGTAAAAAAGTTGTGCTGCTCTCTGCGACGTAACGCGAAAGATGAACGCGTGCTTTTTCATTACAACGGACACGGCGTACCCAAGCCGACGGCGCAGGGAGAGATCTGGGTCTTTAATAGGACTTACACCCAGTACATCCCGCTGTCGATGTACGACCTGCAGACGTGGATGGGCGCGCCCTCGCTCTACGTGTACGACTGCTCCAACGCCGGCATCATTGTCGACAGCTTCAAGTTGTTTGCTGATCAACACGAGAGGGAGTACGAGGCTCAGAAGAATGAAGAAGGCACTTCGAGCATCCCTCAGGTCTCGTACAAGAACTGCATCCAGCTGGCCGCCTGCGCCGCCGGACAGAGCCTGCCCATGAACCCGGAGCTGCCCGCGGACCTGTTCACGGCCTGCCTCACCACGCCGGTCAAGACTGCCATGAAGTGGTTCGTGCTGAGGACCCGGACCAGATACGCCGACAGGGACCTGCACGACCTCATCGATAAGATCCCTGGCCAAGTGACGGACCGCCGCACCATGCTGGGCGAACTGAACTGGATCTTCACGGCCATCACCGACACCATCGCCTGGAGCTCGCTGCCCGCCGACCTCTTCCAGCAGCTGTTCAGGGCCGACTTGCTGACGGCCAGCCTCTGCAGGAACTTCTTACTCGCCGACAGGATTATGAGATCATACAACTGCACCCCCGTGAGCAGTCCGGCGCTGCCGTCACTGGCCGCGCACCCGCTGTGGGCGGCGTGGGAGCAGACCCTGGACTCGGCGCTGGCCCAGCTGGTCACGTTATCTTCGGCTCCACATCCGCTGGATTACTCCCACTCGCCGTTCTTCAGGGATCAGCTCACCGCCTTCCAAGTCTGGCTAGATCTCGGTAAATGGTGCGGCGGGCGCGGCGTGCCGGAGCAGCTGCCGATGGTGCTGCAGGTGCTGCTGAGCACGCTGCACCGGGTGCGCGCGCTGCAGCTGCTGTGCCGGTTCCTGGCGCTGGGCGCGTGGGCGGTGCGCGCCATGCTGGCCGTCGGGATATTCCCCTACATGCTCAAGCTGCTGCAGGCCTCCGCGCCGGTGCTGCGCGCCTCCATGGTCTACATCTGGGCCAGGATCATCGCGGTCGATCCGAGCTGTCAAGCGGACCTAGTCAACGCTAAAGGGCACACGTACTTCCTGTCCGTACTGCAAGACCCTACAGTCGACAGCGAGCACCGCACCCTGGCCGCCTACGTGCTGGCTGGCATCGTGGACGGCTACCCGGCGGGGCAGGAGGCCGCGCTGCAGGGCTCAATGATATCGATATGCCTCGAGCAGCTCGGCGACAGGGAGCCGGTACTGGCCCAGTGGATCTGCATCAGCCTGGGCCGGCTGTGGCGCGGGTTCGAGGCGGCGCGATGGACGGGAGTCAGAGATCTCGCGCACGAGAAACTGTTCCCGCTCCTTGAGCATTCTGAGCCTGAGGTCCGCGCGGCGTGCGCCTTCGCGCTGGGCTCGTTCGTGTGGTGGTGCGGCGGCGCCGGGCGCTGCGAGCAGGCCAACGCGCTGGACCTGCAGGTGGGCGTGCAGCTGGCCGCGCGCCTGCCGCTCGACGCCAGCCCGCTCGTGCGCGCAGAGATACTGGCCGCATTACAGTGGCTGGTGTTAATATTTGAACAACACTTCATCGCTGTTTACATACAAGAACGGGTCAGAAGAAATGATAGGGGTGGCCCGTCCGGGCGCGTGGAGGCGGGGCAGGAGGCGCTGGCGGGGGGCGGCGGGGGCGCGGCGGGAGGTCGGTCCCGGCCCCCCCACTACTCGCTGCCGGCCATCGGGTTCGGCTCCGTGCCGGTCCGGCTGTTCTCCACGCTGTGCGCCATGGCCCGGGAGCCGCACCCCAGGGTCGCGCAGATGGCCACTGACATCCTCAATTACATCTCGAACCAGAGCGCCCAAGTGGACAGCGTGGCGGAACCCGCGCGGAACTCCGCCAGCCTGCCGCCCTCGCCCGGGACCCGCCACCACGAGCACGCCCGGACCCTGCCCGGACACGGCCATCGTCGCGGCAAGTCGGGGCTGCCACACACCATATCCGAGGAGTCTGTTGCGAACAGAGACCGCGACTCTGGAGGGGACCGAGAACGGCACAAAGATTCACTCTCATCTAATTCTAGTCAGACATGTACGAGCAAGAAGGCGATAGTGAGCACCGAGTTCGTGGAGTGGGCGGGCGGCGCGTTCTCCCGGCCCGAGCCCGCGCGCCCGGACTGCGAGAGCCGCGAGCACCACCAGCGCCGCTGGACCCTCGCGCGCAACCGCGAGCTGCGCAGGGAGGCCAGAGCCGCGTGCCGCGTGTCCCGGCTGGAGACGCAGGCCTTCCACTCGCGCTGCCCGCTGCCGCCCGCCGTGGTGCTCTTCCACCCCTACGACCAGCATCTGGCCGTCGCCTCCAAGGATAACTTCGGTATCTGGGACTGGGGCACGGCGGCCAAGCTGTGCGTGGGCTCGTGGCGCCCGGCGTGGGGCCGCATCTCCTCGCTGGCCTACCTCAACGAGCACCAGCACGCGCTGCTGGCCGTGGCCTCGCACACCGGCAACCTGGCAGTCTACAGGCCGTCGGGCAGCAGCACGGAGCCGGCGCTGGTGAGCGCGTGGCGCGCGCTGGACGTGTCCCCCCCGCCGCAGTACACCCCGCCGCGCGCGCAGCCCATCTACAGTATAGCTCAACTGGTCTCTGATCAGTTCTCCTCGAACGAAGACAAACTAGCGACGAACAAAAACAAGCAACAGGCCGACCCCAACTTGGGCGGCGGCACGGTGGTACGCTGGGTGGCGGCGCGGCGCTGGGTGGCGGCGGGCGGGCGCGCGCGCGGCGTGCGGCTGTGGGACGCGCGGCGCGAGCTGCGGGTGGGCGACCTGGCCGCGCCCGGGGACGCCGCCCTCACCGCGCTGGGGGCCGCGCGCCCCGACGTGCTGGCCGCCGGCTTCCAGGACGGCGCCGTGCGGCTGTGGGACCAGCGCACCCTGGCGCCCACGCACGACCTGCGCCGCCACGCCGCGCCCGTGCTGGCCGCCGCGCACTCGCTGCGCGCCGACCTGCTCGTCACCGGCTGCGCGAACGGCGAGATATGTATATACGACACCCGCAAGATGGCCGTGCTGGAGGAAATCCGCGCGCCCGGGCCGCTCGCCGCCGTGGACGTGCACCCGCTGTGCGACCTCGTGGCCTGTGGCTCGCTGAACCAGTGCATCAGCCTGTACGACCTGCGGGGGAACGCGCTCAACACCATCAAGTTCCACGAGGGCTTCATGGGGCCCCGCATCGGGCCCGTCTCCTGTCTGGCCTTCCACCCGCTCAGGTGCGCGATGGGCGTGGGCTCCAAGGACTCCACGGTGTCGGTGTACGTGGCGGAGTCGCGCCGGTGA
Protein
MTEMPPYHAVKDEEKTSGQSDSDSSSDTDDWDLSQCFDKPQHLEPIKGAERVEHSWRVKEKYKTHCVALVLCLNVGVDPPDVVKTQPCARLECWIDPLSLSPSKALESIGHALQSQYERWQPRARYKQSLDPTSEEVKKLCCSLRRNAKDERVLFHYNGHGVPKPTAQGEIWVFNRTYTQYIPLSMYDLQTWMGAPSLYVYDCSNAGIIVDSFKLFADQHEREYEAQKNEEGTSSIPQVSYKNCIQLAACAAGQSLPMNPELPADLFTACLTTPVKTAMKWFVLRTRTRYADRDLHDLIDKIPGQVTDRRTMLGELNWIFTAITDTIAWSSLPADLFQQLFRADLLTASLCRNFLLADRIMRSYNCTPVSSPALPSLAAHPLWAAWEQTLDSALAQLVTLSSAPHPLDYSHSPFFRDQLTAFQVWLDLGKWCGGRGVPEQLPMVLQVLLSTLHRVRALQLLCRFLALGAWAVRAMLAVGIFPYMLKLLQASAPVLRASMVYIWARIIAVDPSCQADLVNAKGHTYFLSVLQDPTVDSEHRTLAAYVLAGIVDGYPAGQEAALQGSMISICLEQLGDREPVLAQWICISLGRLWRGFEAARWTGVRDLAHEKLFPLLEHSEPEVRAACAFALGSFVWWCGGAGRCEQANALDLQVGVQLAARLPLDASPLVRAEILAALQWLVLIFEQHFIAVYIQERVRRNDRGGPSGRVEAGQEALAGGGGGAAGGRSRPPHYSLPAIGFGSVPVRLFSTLCAMAREPHPRVAQMATDILNYISNQSAQVDSVAEPARNSASLPPSPGTRHHEHARTLPGHGHRRGKSGLPHTISEESVANRDRDSGGDRERHKDSLSSNSSQTCTSKKAIVSTEFVEWAGGAFSRPEPARPDCESREHHQRRWTLARNRELRREARAACRVSRLETQAFHSRCPLPPAVVLFHPYDQHLAVASKDNFGIWDWGTAAKLCVGSWRPAWGRISSLAYLNEHQHALLAVASHTGNLAVYRPSGSSTEPALVSAWRALDVSPPPQYTPPRAQPIYSIAQLVSDQFSSNEDKLATNKNKQQADPNLGGGTVVRWVAARRWVAAGGRARGVRLWDARRELRVGDLAAPGDAALTALGAARPDVLAAGFQDGAVRLWDQRTLAPTHDLRRHAAPVLAAAHSLRADLLVTGCANGEICIYDTRKMAVLEEIRAPGPLAAVDVHPLCDLVACGSLNQCISLYDLRGNALNTIKFHEGFMGPRIGPVSCLAFHPLRCAMGVGSKDSTVSVYVAESRR
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
D6N9W7
H9J7N5
A0A212FF82
A0A0C9QP95
E0VU38
D6WXI0
+ More
E2ASD4 F4WBK8 E2BZ70 A0A195C1Q1 A0A158NNY1 A0A0M4M1W1 A0A195FS11 A0A151X801 A0A1W4WNJ0 A0A151HZI5 A0A195DAY8 N6SZU7 T1IDV6 A0A0A9WPK3 A0A0J7KL19 U5ETP3 A0A232F979 A0A1B0CH83 A0A1Y1KRJ0 A0A1Q3F143 A0A0M8ZX93 A0A2A3E2U7 A0A3S3Q0Y0 A0A088A8W7 A0A3L8DEC7 A0A2L2YCX6 A0A0P5YZ31 A0A0P5J420 A0A1B6L3U9 A0A0P6FAX8 A0A0P5IJP5 A0A0P5SHU1 A0A0P5NYK1 A0A1L8DX75 A0A0P5IR88 A0A1I8M9W8 A0A1A9WAX1 A0A1B6F1B6 A0A1A9VUH8 A0A1B0GB21 A0A1A9XLR4 A0A1B0AC27 A0A1B0BKD2 A0A0P6FBP0 A0A1D1W1X9 A0A1L8DWZ7 T1PPJ9 K1PCZ2 A0A182IJQ6 A0A182SR47 A0A1I8PYD5 A0A0P5J8I1 A0A0P4ZYK9 A0A182QVZ5
E2ASD4 F4WBK8 E2BZ70 A0A195C1Q1 A0A158NNY1 A0A0M4M1W1 A0A195FS11 A0A151X801 A0A1W4WNJ0 A0A151HZI5 A0A195DAY8 N6SZU7 T1IDV6 A0A0A9WPK3 A0A0J7KL19 U5ETP3 A0A232F979 A0A1B0CH83 A0A1Y1KRJ0 A0A1Q3F143 A0A0M8ZX93 A0A2A3E2U7 A0A3S3Q0Y0 A0A088A8W7 A0A3L8DEC7 A0A2L2YCX6 A0A0P5YZ31 A0A0P5J420 A0A1B6L3U9 A0A0P6FAX8 A0A0P5IJP5 A0A0P5SHU1 A0A0P5NYK1 A0A1L8DX75 A0A0P5IR88 A0A1I8M9W8 A0A1A9WAX1 A0A1B6F1B6 A0A1A9VUH8 A0A1B0GB21 A0A1A9XLR4 A0A1B0AC27 A0A1B0BKD2 A0A0P6FBP0 A0A1D1W1X9 A0A1L8DWZ7 T1PPJ9 K1PCZ2 A0A182IJQ6 A0A182SR47 A0A1I8PYD5 A0A0P5J8I1 A0A0P4ZYK9 A0A182QVZ5
Pubmed
EMBL
GU350774
ADB91965.1
BABH01020429
BABH01020430
BABH01020431
BABH01020432
+ More
BABH01020433 AGBW02008849 OWR52387.1 GBYB01005399 JAG75166.1 DS235779 EEB16894.1 KQ971361 EFA08847.1 GL442284 EFN63668.1 GL888065 EGI68413.1 GL451577 EFN78976.1 KQ978344 KYM94804.1 ADTU01002780 KR075849 ALE20568.1 KQ981285 KYN43216.1 KQ982431 KYQ56454.1 KQ976693 KYM77649.1 KQ981042 KYN10053.1 APGK01057803 KB741282 KB631802 ENN70788.1 ERL86240.1 ACPB03000196 GBHO01033930 JAG09674.1 LBMM01005959 KMQ91063.1 GANO01004283 JAB55588.1 NNAY01000614 OXU27386.1 AJWK01012105 AJWK01012106 AJWK01012107 GEZM01078063 JAV63031.1 GFDL01013796 JAV21249.1 KQ435812 KOX72777.1 KZ288467 PBC25421.1 NCKU01001580 NCKU01001553 RWS11751.1 RWS11827.1 QOIP01000009 RLU18249.1 IAAA01019167 LAA05035.1 GDIP01051187 JAM52528.1 GDIQ01205533 JAK46192.1 GEBQ01021584 JAT18393.1 GDIQ01066087 JAN28650.1 GDIQ01213984 JAK37741.1 GDIQ01093170 GDIQ01066088 JAL58556.1 GDIQ01138017 JAL13709.1 GFDF01003065 JAV11019.1 GDIQ01211733 JAK39992.1 GECZ01025822 JAS43947.1 CCAG010006530 JXJN01015893 GDIQ01052494 JAN42243.1 BDGG01000013 GAV06118.1 GFDF01003145 JAV10939.1 KA650110 AFP64739.1 JH818602 EKC19393.1 GDIQ01228578 JAK23147.1 GDIP01206814 JAJ16588.1 AXCN02002096 AXCN02002097
BABH01020433 AGBW02008849 OWR52387.1 GBYB01005399 JAG75166.1 DS235779 EEB16894.1 KQ971361 EFA08847.1 GL442284 EFN63668.1 GL888065 EGI68413.1 GL451577 EFN78976.1 KQ978344 KYM94804.1 ADTU01002780 KR075849 ALE20568.1 KQ981285 KYN43216.1 KQ982431 KYQ56454.1 KQ976693 KYM77649.1 KQ981042 KYN10053.1 APGK01057803 KB741282 KB631802 ENN70788.1 ERL86240.1 ACPB03000196 GBHO01033930 JAG09674.1 LBMM01005959 KMQ91063.1 GANO01004283 JAB55588.1 NNAY01000614 OXU27386.1 AJWK01012105 AJWK01012106 AJWK01012107 GEZM01078063 JAV63031.1 GFDL01013796 JAV21249.1 KQ435812 KOX72777.1 KZ288467 PBC25421.1 NCKU01001580 NCKU01001553 RWS11751.1 RWS11827.1 QOIP01000009 RLU18249.1 IAAA01019167 LAA05035.1 GDIP01051187 JAM52528.1 GDIQ01205533 JAK46192.1 GEBQ01021584 JAT18393.1 GDIQ01066087 JAN28650.1 GDIQ01213984 JAK37741.1 GDIQ01093170 GDIQ01066088 JAL58556.1 GDIQ01138017 JAL13709.1 GFDF01003065 JAV11019.1 GDIQ01211733 JAK39992.1 GECZ01025822 JAS43947.1 CCAG010006530 JXJN01015893 GDIQ01052494 JAN42243.1 BDGG01000013 GAV06118.1 GFDF01003145 JAV10939.1 KA650110 AFP64739.1 JH818602 EKC19393.1 GDIQ01228578 JAK23147.1 GDIP01206814 JAJ16588.1 AXCN02002096 AXCN02002097
Proteomes
UP000005204
UP000007151
UP000009046
UP000007266
UP000000311
UP000007755
+ More
UP000008237 UP000078542 UP000005205 UP000078541 UP000075809 UP000192223 UP000078540 UP000078492 UP000019118 UP000030742 UP000015103 UP000036403 UP000215335 UP000092461 UP000053105 UP000242457 UP000285301 UP000005203 UP000279307 UP000095301 UP000091820 UP000078200 UP000092444 UP000092443 UP000092445 UP000092460 UP000186922 UP000005408 UP000075880 UP000075901 UP000095300 UP000075886
UP000008237 UP000078542 UP000005205 UP000078541 UP000075809 UP000192223 UP000078540 UP000078492 UP000019118 UP000030742 UP000015103 UP000036403 UP000215335 UP000092461 UP000053105 UP000242457 UP000285301 UP000005203 UP000279307 UP000095301 UP000091820 UP000078200 UP000092444 UP000092443 UP000092445 UP000092460 UP000186922 UP000005408 UP000075880 UP000075901 UP000095300 UP000075886
Interpro
IPR016024
ARM-type_fold
+ More
IPR001680 WD40_repeat
IPR004083 Raptor
IPR029347 Raptor_N
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR011989 ARM-like
IPR000357 HEAT
IPR024977 Apc4_WD40_dom
IPR019775 WD40_repeat_CS
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR011047 Quinoprotein_ADH-like_supfam
IPR001680 WD40_repeat
IPR004083 Raptor
IPR029347 Raptor_N
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR011989 ARM-like
IPR000357 HEAT
IPR024977 Apc4_WD40_dom
IPR019775 WD40_repeat_CS
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR011047 Quinoprotein_ADH-like_supfam
Gene 3D
ProteinModelPortal
D6N9W7
H9J7N5
A0A212FF82
A0A0C9QP95
E0VU38
D6WXI0
+ More
E2ASD4 F4WBK8 E2BZ70 A0A195C1Q1 A0A158NNY1 A0A0M4M1W1 A0A195FS11 A0A151X801 A0A1W4WNJ0 A0A151HZI5 A0A195DAY8 N6SZU7 T1IDV6 A0A0A9WPK3 A0A0J7KL19 U5ETP3 A0A232F979 A0A1B0CH83 A0A1Y1KRJ0 A0A1Q3F143 A0A0M8ZX93 A0A2A3E2U7 A0A3S3Q0Y0 A0A088A8W7 A0A3L8DEC7 A0A2L2YCX6 A0A0P5YZ31 A0A0P5J420 A0A1B6L3U9 A0A0P6FAX8 A0A0P5IJP5 A0A0P5SHU1 A0A0P5NYK1 A0A1L8DX75 A0A0P5IR88 A0A1I8M9W8 A0A1A9WAX1 A0A1B6F1B6 A0A1A9VUH8 A0A1B0GB21 A0A1A9XLR4 A0A1B0AC27 A0A1B0BKD2 A0A0P6FBP0 A0A1D1W1X9 A0A1L8DWZ7 T1PPJ9 K1PCZ2 A0A182IJQ6 A0A182SR47 A0A1I8PYD5 A0A0P5J8I1 A0A0P4ZYK9 A0A182QVZ5
E2ASD4 F4WBK8 E2BZ70 A0A195C1Q1 A0A158NNY1 A0A0M4M1W1 A0A195FS11 A0A151X801 A0A1W4WNJ0 A0A151HZI5 A0A195DAY8 N6SZU7 T1IDV6 A0A0A9WPK3 A0A0J7KL19 U5ETP3 A0A232F979 A0A1B0CH83 A0A1Y1KRJ0 A0A1Q3F143 A0A0M8ZX93 A0A2A3E2U7 A0A3S3Q0Y0 A0A088A8W7 A0A3L8DEC7 A0A2L2YCX6 A0A0P5YZ31 A0A0P5J420 A0A1B6L3U9 A0A0P6FAX8 A0A0P5IJP5 A0A0P5SHU1 A0A0P5NYK1 A0A1L8DX75 A0A0P5IR88 A0A1I8M9W8 A0A1A9WAX1 A0A1B6F1B6 A0A1A9VUH8 A0A1B0GB21 A0A1A9XLR4 A0A1B0AC27 A0A1B0BKD2 A0A0P6FBP0 A0A1D1W1X9 A0A1L8DWZ7 T1PPJ9 K1PCZ2 A0A182IJQ6 A0A182SR47 A0A1I8PYD5 A0A0P5J8I1 A0A0P4ZYK9 A0A182QVZ5
PDB
6BCX
E-value=0,
Score=2142
Ontologies
PATHWAY
GO
PANTHER
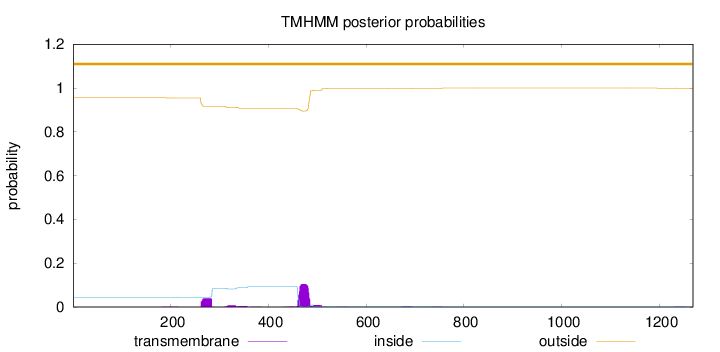
Topology
Length:
1269
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.70028
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04439
outside
1 - 1269
Population Genetic Test Statistics
Pi
244.901311
Theta
175.113181
Tajima's D
2.036242
CLR
0.250496
CSRT
0.88135593220339
Interpretation
Uncertain
