Gene
KWMTBOMO10038
Pre Gene Modal
BGIBMGA005526
Annotation
putative_light_protein?_partial_[Operophtera_brumata]
Full name
Vacuolar protein sorting-associated protein 41 homolog
Location in the cell
Cytoplasmic Reliability : 2.348
Sequence
CDS
ATGGCTGTTAATATTGAGAGCTGCGAAAGCTTGCCTCCCGATGAGGAGCCGAAGCTAAAATACGATAGAATGGAAAATGATTTATTGAATATTTTACTGAAAGACGCTGTCAGTTGTATTTGTGTTCACACCAAATTCATCTGTATCGGAACACGTTGGGGTGTGATACACCTGCTAGATCACGAAGGTAACATAGTTGCTATATCCCAGGACGGGAAGACTGAATTCCAGGCTCATACTATCGCTGTCAACCAAATATCTGTAGACCAAAATGGGGATTACATAGCGAGTTGTTCAGACGATGGTAAAGTTGTCGTATATGGATTGTACAGTAATGATAACACGCACAACCTAAATTTAGGCAGGGCAGTGAAATCAGTTGCATTGGATCCTTGCTACTTCAAGTCAGGTTCAGGAAGAAGATTTCTTACAGGTGATCACAGATTGACACTATATGAAAAATCATTCCTTAATCGAATACGAACTACAGTATTACATGAATGCAAGGGAAACGTACAAGCCATGTCCTGGCAAGACAGGTTTGTAGCATGGGCAAGTGAAGTTGGGGTCCGTGTTTATGACCTAGTTGCTAAATGCTCTCTAGGTCTCATCCAATGGGAAAAGAACCTAAATAGATCTATTGAAGACTACCGCTGCAATCTTCTCTGGTCTGCTCCTAAGACTCTAATGATCGGCTGGGTTGATACCATCAGGATATGTGTCATTAGAAAGCGAAGTCAAATTGAATTACAGACTCGTGATGTCACAGAATACCTCGTGGATCCAATATATACCTACCAAATAGATTATTTTATAGGAGGTCTGGGCCCTTTGGACGATCTATTAGTTTTATTAGCCGTTCCAAAAGAATGCGATCCTAAAACCGGAAAGTCGCAAATGCCCGTTTTAATAGTGGCGAATTATAAAGGCTGCGAATTCTGCGAGATATCTTGGGAGACTTTAGACATAAGGGGCTACCAAGAATATAACTGTAATGATTACCATTTAGATATGCTTATAGAAGAAAACAGATTTTTTATTGTTTCCCCCAAAGACATTATCATAGCGAGCCCTTACGACATTGACGATAGAGTCACTTGGTTGACCCAACATAGAAGGTTCGAAAAAGCTATTGCCGTGTTGGAAGAGGTCGGTGGTAAAACTTCTAAGCACTCCGTGATGACGGTGGGCGTACAGTATTTAGATCACTTGCTCTCTGAACATCTTTATCAAGAGGCTGCCATTCTCTGTGCCAGAATATTTAAAAACGATAAAGTTTTATTAGAAAATCAAATATTAAAGTTTGCCGGCGTCAAACAACTTAGAACTATAAGTCCCTACGTGCCGAAGACGCCTGAACAAGCTTTAAGTCCTTACATATACGAAATAATACTTTTCGAATACCTCGAAGTGGATTCGCGAGGATTTTTAAAAATAATTAAAGAATGGAATCCAACTCTATACAAAACGAGCGTCGTTATAAACAAAGTAATCGAACATTTGTTAAACACCGAAGTTGACACGAACGTGTATCTGGAAACCTTAGCTTTACTTTACTGCCACCAAAAGCAATACGATAAAGCGCTGACAACATTCTTAACATTGCAACACAAGGATGTATTCAAATTGATAACAGACCACGAAATCTACAACATCATACAGGACAAGATACTCGATCTAATGACGCTAGACTGCGACCAGGCTATATCGATACTCTTGAATAACACAAAAATCCCTGTTGAGGTCGTCGAACAACAACTGGCAAATCACGAAGAATATTTGTATAAGTACTTAGACGCGTACAGCAAGATGGAACCGAACGGTCGATACCATGGAAAACTGGTTCGTCTATACGCCAAATACAACAAAGAAAAACTGTTGCCTTTCTTAAGATGCAGCGATAATTATCCCATACAGGAAGCTTTAGACGTGTGTCAGGCCAACAATTTTTTCCCAGAAATGGTTTTTCTACTCGCGAGAATCGGCAACACCAAGGAGGCATTGCAGATAATTATCGAACAGTTAAACGACATAAACCAAGCCATAACTTTCTGTCAAGAACAAAACGATATGGAACTTTGGATAGATCTGATACAGCAGACGGTCGACAAGCCGGAATATGTCACTTTACTTCTGAAAAGGATAGGGAACTACGTGGACCCGAGGATGTTGATACAGAACATCAAACCGGGCTGCGAAATAAAAGACCTCAAAGAGTGCTTAGCCAAAATGATGTGCGACTATCATCTCCAGCTATCAGTGCAGGAGGCCTGTAAAGTCATTACGCTGAGGAACTATTTTGATTTGCATGAAAAGCAAGTGATGGCTCAACAACGAGGCATATCGGTGACCGACGACTTCCTCTGCTGCGTTTGTCACGGGCGGATCATAATAAGGGACATGTCGAATGCTTCTGACCTTCTAGTTTACAATTGCAGACATTCCTTCCACGCCGGCTGCCTCCCGGAAGGCGACCAGTGTGCTGTTTGTAATGCGGTCAGAATGTGA
Protein
MAVNIESCESLPPDEEPKLKYDRMENDLLNILLKDAVSCICVHTKFICIGTRWGVIHLLDHEGNIVAISQDGKTEFQAHTIAVNQISVDQNGDYIASCSDDGKVVVYGLYSNDNTHNLNLGRAVKSVALDPCYFKSGSGRRFLTGDHRLTLYEKSFLNRIRTTVLHECKGNVQAMSWQDRFVAWASEVGVRVYDLVAKCSLGLIQWEKNLNRSIEDYRCNLLWSAPKTLMIGWVDTIRICVIRKRSQIELQTRDVTEYLVDPIYTYQIDYFIGGLGPLDDLLVLLAVPKECDPKTGKSQMPVLIVANYKGCEFCEISWETLDIRGYQEYNCNDYHLDMLIEENRFFIVSPKDIIIASPYDIDDRVTWLTQHRRFEKAIAVLEEVGGKTSKHSVMTVGVQYLDHLLSEHLYQEAAILCARIFKNDKVLLENQILKFAGVKQLRTISPYVPKTPEQALSPYIYEIILFEYLEVDSRGFLKIIKEWNPTLYKTSVVINKVIEHLLNTEVDTNVYLETLALLYCHQKQYDKALTTFLTLQHKDVFKLITDHEIYNIIQDKILDLMTLDCDQAISILLNNTKIPVEVVEQQLANHEEYLYKYLDAYSKMEPNGRYHGKLVRLYAKYNKEKLLPFLRCSDNYPIQEALDVCQANNFFPEMVFLLARIGNTKEALQIIIEQLNDINQAITFCQEQNDMELWIDLIQQTVDKPEYVTLLLKRIGNYVDPRMLIQNIKPGCEIKDLKECLAKMMCDYHLQLSVQEACKVITLRNYFDLHEKQVMAQQRGISVTDDFLCCVCHGRIIIRDMSNASDLLVYNCRHSFHAGCLPEGDQCAVCNAVRM
Summary
Description
Plays a role in vesicle-mediated protein trafficking to lysosomal compartments including the endocytic membrane transport pathways.
Similarity
Belongs to the VPS41 family.
Belongs to the FBPase class 1 family.
Belongs to the FBPase class 1 family.
Uniprot
H9J7N4
A0A2A4JGH8
A0A2W1BI69
A0A2H1WTJ1
A0A0L7LRU4
A0A212FF77
+ More
A0A194QAH7 A0A0N1IPT3 A0A3S2PEC0 A0A067QY06 A0A1Y1M1I5 D2A5A3 A0A088AMB9 A0A2A3E7V0 E2BHH5 A0A232EXD2 E2ABX3 A0A0M9AA07 A0A1L8DVT6 A0A158NL78 A0A195BY12 A0A195BFV3 A0A1Q3FE53 A0A151JSM3 A0A151WHF1 A0A195F0P5 A0A0C9PHG4 B0WUB3 A0A084VMX0 W5JRN4 A0A2M4CY22 A0A2M4AD66 A0A2M4ACI9 A0A0N8ERW9 A0A1I8JSK6 A0A182Y9S3 A0A2J7Q9R1 A0A182IX16 A0A182QF86 A0A182M9Z9 A0A182WH67 A0A182PFR5 A0A182R6J0 A0A182KAX8 A0A182VF10 A0A1I8JVZ5 A0A1Y9GKU3 Q7PVV4 E9J123 U4UYK7 N6TLU3 A0A1S4H0D6 A0A3L8DTA4 F4X642 A0A2M4ADT6 A0A310SC13 A0A182NF47 A0A336KCB0 A0A1B6BZ97 A0A1B6D753 A0A1B6CQD6 A0A1B6C2S4 A0A182UJC0 A0A0P6I3T2 A0A0P5D2G6 A0A0P5BQ39 A0A0P5XIT6 A0A162CAG7 A0A0N8BYH2 A0A0P6FXI0 A0A0L7RCU8 E9FYK6 J9K6C7 A0A0P4Y1X2 A0A0P6F0Y2 A0A154P1E0 A0A2H8TUJ9 A0A0K8TMJ4 A0A0N8C5S3 A0A0P5D0K8 A0A0N7ZKY4 A0A224X744 A0A069DWL6 E0VK08 A0A0P5PYV0 A0A023F2M8 A0A0P4W4B1 A0A0K8VL42 A0A1A9WES6 A0A1B0FA38 B4KFT8 A0A1I8M5E1 A0A1I8PEM5 A0A0A1WFZ4 A0A0P5DRT4 A0A0P5VB79 A0A0A9XUE9 A0A0L0C426 A0A0P4YYI3 A0A3B0JYN3
A0A194QAH7 A0A0N1IPT3 A0A3S2PEC0 A0A067QY06 A0A1Y1M1I5 D2A5A3 A0A088AMB9 A0A2A3E7V0 E2BHH5 A0A232EXD2 E2ABX3 A0A0M9AA07 A0A1L8DVT6 A0A158NL78 A0A195BY12 A0A195BFV3 A0A1Q3FE53 A0A151JSM3 A0A151WHF1 A0A195F0P5 A0A0C9PHG4 B0WUB3 A0A084VMX0 W5JRN4 A0A2M4CY22 A0A2M4AD66 A0A2M4ACI9 A0A0N8ERW9 A0A1I8JSK6 A0A182Y9S3 A0A2J7Q9R1 A0A182IX16 A0A182QF86 A0A182M9Z9 A0A182WH67 A0A182PFR5 A0A182R6J0 A0A182KAX8 A0A182VF10 A0A1I8JVZ5 A0A1Y9GKU3 Q7PVV4 E9J123 U4UYK7 N6TLU3 A0A1S4H0D6 A0A3L8DTA4 F4X642 A0A2M4ADT6 A0A310SC13 A0A182NF47 A0A336KCB0 A0A1B6BZ97 A0A1B6D753 A0A1B6CQD6 A0A1B6C2S4 A0A182UJC0 A0A0P6I3T2 A0A0P5D2G6 A0A0P5BQ39 A0A0P5XIT6 A0A162CAG7 A0A0N8BYH2 A0A0P6FXI0 A0A0L7RCU8 E9FYK6 J9K6C7 A0A0P4Y1X2 A0A0P6F0Y2 A0A154P1E0 A0A2H8TUJ9 A0A0K8TMJ4 A0A0N8C5S3 A0A0P5D0K8 A0A0N7ZKY4 A0A224X744 A0A069DWL6 E0VK08 A0A0P5PYV0 A0A023F2M8 A0A0P4W4B1 A0A0K8VL42 A0A1A9WES6 A0A1B0FA38 B4KFT8 A0A1I8M5E1 A0A1I8PEM5 A0A0A1WFZ4 A0A0P5DRT4 A0A0P5VB79 A0A0A9XUE9 A0A0L0C426 A0A0P4YYI3 A0A3B0JYN3
Pubmed
19121390
28756777
26227816
22118469
26354079
24845553
+ More
28004739 18362917 19820115 20798317 28648823 21347285 24438588 20920257 23761445 26999592 25244985 12364791 21282665 23537049 30249741 21719571 21292972 26369729 26334808 20566863 25474469 27129103 17994087 25315136 25830018 25401762 26823975 26108605
28004739 18362917 19820115 20798317 28648823 21347285 24438588 20920257 23761445 26999592 25244985 12364791 21282665 23537049 30249741 21719571 21292972 26369729 26334808 20566863 25474469 27129103 17994087 25315136 25830018 25401762 26823975 26108605
EMBL
BABH01020434
NWSH01001549
PCG70886.1
KZ150179
PZC72526.1
ODYU01010946
+ More
SOQ56371.1 JTDY01000229 KOB78185.1 AGBW02008849 OWR52386.1 KQ459232 KPJ02538.1 KQ460044 KPJ18302.1 RSAL01000075 RVE48905.1 KK853123 KDR11061.1 GEZM01043149 JAV79271.1 KQ971345 EFA05104.1 KZ288340 PBC27770.1 GL448301 EFN84828.1 NNAY01001745 OXU23046.1 GL438386 EFN69068.1 KQ435711 KOX79536.1 GFDF01003544 JAV10540.1 ADTU01019279 ADTU01019280 ADTU01019281 ADTU01019282 KQ978501 KYM93549.1 KQ976488 KYM83490.1 GFDL01009211 JAV25834.1 KQ978530 KYN30284.1 KQ983120 KYQ47238.1 KQ981866 KYN34048.1 GBYB01000263 JAG70030.1 DS232103 EDS34885.1 ATLV01014632 KE524975 KFB39314.1 ADMH02000666 ETN65394.1 GGFL01006054 MBW70232.1 GGFK01005351 MBW38672.1 GGFK01005176 MBW38497.1 GDUN01001098 JAN94821.1 NEVH01016344 PNF25314.1 AXCN02000829 AXCM01001716 APCN01003788 AAAB01008984 EAA14980.4 GL767540 EFZ13482.1 KB632429 ERL95476.1 APGK01019082 KB740092 ENN81419.1 QOIP01000004 RLU23670.1 GL888769 EGI58075.1 GGFK01005633 MBW38954.1 KQ761619 OAD57320.1 UFQS01000259 UFQT01000259 SSX02151.1 SSX22528.1 GEDC01030712 JAS06586.1 GEDC01015781 JAS21517.1 GEDC01021677 JAS15621.1 GEDC01029487 JAS07811.1 GDIQ01010009 JAN84728.1 GDIP01161982 JAJ61420.1 GDIP01186271 JAJ37131.1 GDIP01071491 JAM32224.1 LRGB01001151 KZS13472.1 GDIQ01132483 JAL19243.1 GDIQ01052703 JAN42034.1 KQ414615 KOC68581.1 GL732527 EFX87545.1 ABLF02040091 ABLF02040104 ABLF02040105 ABLF02040112 GDIP01233711 JAI89690.1 GDIQ01054544 JAN40193.1 KQ434796 KZC05745.1 GFXV01005043 MBW16848.1 GDAI01002232 JAI15371.1 GDIQ01112075 JAL39651.1 GDIP01164538 JAJ58864.1 GDIP01235265 JAI88136.1 GFTR01008149 JAW08277.1 GBGD01000456 JAC88433.1 DS235235 EEB13714.1 GDIQ01123626 JAL28100.1 GBBI01003331 JAC15381.1 GDKW01000417 JAI56178.1 GDHF01013059 JAI39255.1 CCAG010005354 CH933807 EDW12064.1 GBXI01016338 JAC97953.1 GDIP01151568 JAJ71834.1 GDIP01102726 JAM00989.1 GBHO01021076 GBHO01021074 GBRD01015152 GDHC01012615 JAG22528.1 JAG22530.1 JAG50674.1 JAQ06014.1 JRES01000940 KNC26991.1 GDIP01220763 JAJ02639.1 OUUW01000010 SPP85542.1
SOQ56371.1 JTDY01000229 KOB78185.1 AGBW02008849 OWR52386.1 KQ459232 KPJ02538.1 KQ460044 KPJ18302.1 RSAL01000075 RVE48905.1 KK853123 KDR11061.1 GEZM01043149 JAV79271.1 KQ971345 EFA05104.1 KZ288340 PBC27770.1 GL448301 EFN84828.1 NNAY01001745 OXU23046.1 GL438386 EFN69068.1 KQ435711 KOX79536.1 GFDF01003544 JAV10540.1 ADTU01019279 ADTU01019280 ADTU01019281 ADTU01019282 KQ978501 KYM93549.1 KQ976488 KYM83490.1 GFDL01009211 JAV25834.1 KQ978530 KYN30284.1 KQ983120 KYQ47238.1 KQ981866 KYN34048.1 GBYB01000263 JAG70030.1 DS232103 EDS34885.1 ATLV01014632 KE524975 KFB39314.1 ADMH02000666 ETN65394.1 GGFL01006054 MBW70232.1 GGFK01005351 MBW38672.1 GGFK01005176 MBW38497.1 GDUN01001098 JAN94821.1 NEVH01016344 PNF25314.1 AXCN02000829 AXCM01001716 APCN01003788 AAAB01008984 EAA14980.4 GL767540 EFZ13482.1 KB632429 ERL95476.1 APGK01019082 KB740092 ENN81419.1 QOIP01000004 RLU23670.1 GL888769 EGI58075.1 GGFK01005633 MBW38954.1 KQ761619 OAD57320.1 UFQS01000259 UFQT01000259 SSX02151.1 SSX22528.1 GEDC01030712 JAS06586.1 GEDC01015781 JAS21517.1 GEDC01021677 JAS15621.1 GEDC01029487 JAS07811.1 GDIQ01010009 JAN84728.1 GDIP01161982 JAJ61420.1 GDIP01186271 JAJ37131.1 GDIP01071491 JAM32224.1 LRGB01001151 KZS13472.1 GDIQ01132483 JAL19243.1 GDIQ01052703 JAN42034.1 KQ414615 KOC68581.1 GL732527 EFX87545.1 ABLF02040091 ABLF02040104 ABLF02040105 ABLF02040112 GDIP01233711 JAI89690.1 GDIQ01054544 JAN40193.1 KQ434796 KZC05745.1 GFXV01005043 MBW16848.1 GDAI01002232 JAI15371.1 GDIQ01112075 JAL39651.1 GDIP01164538 JAJ58864.1 GDIP01235265 JAI88136.1 GFTR01008149 JAW08277.1 GBGD01000456 JAC88433.1 DS235235 EEB13714.1 GDIQ01123626 JAL28100.1 GBBI01003331 JAC15381.1 GDKW01000417 JAI56178.1 GDHF01013059 JAI39255.1 CCAG010005354 CH933807 EDW12064.1 GBXI01016338 JAC97953.1 GDIP01151568 JAJ71834.1 GDIP01102726 JAM00989.1 GBHO01021076 GBHO01021074 GBRD01015152 GDHC01012615 JAG22528.1 JAG22530.1 JAG50674.1 JAQ06014.1 JRES01000940 KNC26991.1 GDIP01220763 JAJ02639.1 OUUW01000010 SPP85542.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000283053 UP000027135 UP000007266 UP000005203 UP000242457 UP000008237 UP000215335 UP000000311 UP000053105 UP000005205 UP000078542 UP000078540 UP000078492 UP000075809 UP000078541 UP000002320 UP000030765 UP000000673 UP000069272 UP000076408 UP000235965 UP000075880 UP000075886 UP000075883 UP000075920 UP000075885 UP000075900 UP000075881 UP000075903 UP000076407 UP000075840 UP000007062 UP000030742 UP000019118 UP000279307 UP000007755 UP000075884 UP000075902 UP000076858 UP000053825 UP000000305 UP000007819 UP000076502 UP000009046 UP000091820 UP000092444 UP000009192 UP000095301 UP000095300 UP000037069 UP000268350
UP000283053 UP000027135 UP000007266 UP000005203 UP000242457 UP000008237 UP000215335 UP000000311 UP000053105 UP000005205 UP000078542 UP000078540 UP000078492 UP000075809 UP000078541 UP000002320 UP000030765 UP000000673 UP000069272 UP000076408 UP000235965 UP000075880 UP000075886 UP000075883 UP000075920 UP000075885 UP000075900 UP000075881 UP000075903 UP000076407 UP000075840 UP000007062 UP000030742 UP000019118 UP000279307 UP000007755 UP000075884 UP000075902 UP000076858 UP000053825 UP000000305 UP000007819 UP000076502 UP000009046 UP000091820 UP000092444 UP000009192 UP000095301 UP000095300 UP000037069 UP000268350
PRIDE
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR001841 Znf_RING
IPR011990 TPR-like_helical_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR016024 ARM-type_fold
IPR017986 WD40_repeat_dom
IPR000547 Clathrin_H-chain/VPS_repeat
IPR029347 Raptor_N
IPR004083 Raptor
IPR011989 ARM-like
IPR016902 VPS41
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR033391 FBPase_N
IPR000146 FBPase_class-1
IPR020548 Fructose_bisphosphatase_AS
IPR028343 FBPtase
IPR001841 Znf_RING
IPR011990 TPR-like_helical_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR016024 ARM-type_fold
IPR017986 WD40_repeat_dom
IPR000547 Clathrin_H-chain/VPS_repeat
IPR029347 Raptor_N
IPR004083 Raptor
IPR011989 ARM-like
IPR016902 VPS41
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR033391 FBPase_N
IPR000146 FBPase_class-1
IPR020548 Fructose_bisphosphatase_AS
IPR028343 FBPtase
Gene 3D
CDD
ProteinModelPortal
H9J7N4
A0A2A4JGH8
A0A2W1BI69
A0A2H1WTJ1
A0A0L7LRU4
A0A212FF77
+ More
A0A194QAH7 A0A0N1IPT3 A0A3S2PEC0 A0A067QY06 A0A1Y1M1I5 D2A5A3 A0A088AMB9 A0A2A3E7V0 E2BHH5 A0A232EXD2 E2ABX3 A0A0M9AA07 A0A1L8DVT6 A0A158NL78 A0A195BY12 A0A195BFV3 A0A1Q3FE53 A0A151JSM3 A0A151WHF1 A0A195F0P5 A0A0C9PHG4 B0WUB3 A0A084VMX0 W5JRN4 A0A2M4CY22 A0A2M4AD66 A0A2M4ACI9 A0A0N8ERW9 A0A1I8JSK6 A0A182Y9S3 A0A2J7Q9R1 A0A182IX16 A0A182QF86 A0A182M9Z9 A0A182WH67 A0A182PFR5 A0A182R6J0 A0A182KAX8 A0A182VF10 A0A1I8JVZ5 A0A1Y9GKU3 Q7PVV4 E9J123 U4UYK7 N6TLU3 A0A1S4H0D6 A0A3L8DTA4 F4X642 A0A2M4ADT6 A0A310SC13 A0A182NF47 A0A336KCB0 A0A1B6BZ97 A0A1B6D753 A0A1B6CQD6 A0A1B6C2S4 A0A182UJC0 A0A0P6I3T2 A0A0P5D2G6 A0A0P5BQ39 A0A0P5XIT6 A0A162CAG7 A0A0N8BYH2 A0A0P6FXI0 A0A0L7RCU8 E9FYK6 J9K6C7 A0A0P4Y1X2 A0A0P6F0Y2 A0A154P1E0 A0A2H8TUJ9 A0A0K8TMJ4 A0A0N8C5S3 A0A0P5D0K8 A0A0N7ZKY4 A0A224X744 A0A069DWL6 E0VK08 A0A0P5PYV0 A0A023F2M8 A0A0P4W4B1 A0A0K8VL42 A0A1A9WES6 A0A1B0FA38 B4KFT8 A0A1I8M5E1 A0A1I8PEM5 A0A0A1WFZ4 A0A0P5DRT4 A0A0P5VB79 A0A0A9XUE9 A0A0L0C426 A0A0P4YYI3 A0A3B0JYN3
A0A194QAH7 A0A0N1IPT3 A0A3S2PEC0 A0A067QY06 A0A1Y1M1I5 D2A5A3 A0A088AMB9 A0A2A3E7V0 E2BHH5 A0A232EXD2 E2ABX3 A0A0M9AA07 A0A1L8DVT6 A0A158NL78 A0A195BY12 A0A195BFV3 A0A1Q3FE53 A0A151JSM3 A0A151WHF1 A0A195F0P5 A0A0C9PHG4 B0WUB3 A0A084VMX0 W5JRN4 A0A2M4CY22 A0A2M4AD66 A0A2M4ACI9 A0A0N8ERW9 A0A1I8JSK6 A0A182Y9S3 A0A2J7Q9R1 A0A182IX16 A0A182QF86 A0A182M9Z9 A0A182WH67 A0A182PFR5 A0A182R6J0 A0A182KAX8 A0A182VF10 A0A1I8JVZ5 A0A1Y9GKU3 Q7PVV4 E9J123 U4UYK7 N6TLU3 A0A1S4H0D6 A0A3L8DTA4 F4X642 A0A2M4ADT6 A0A310SC13 A0A182NF47 A0A336KCB0 A0A1B6BZ97 A0A1B6D753 A0A1B6CQD6 A0A1B6C2S4 A0A182UJC0 A0A0P6I3T2 A0A0P5D2G6 A0A0P5BQ39 A0A0P5XIT6 A0A162CAG7 A0A0N8BYH2 A0A0P6FXI0 A0A0L7RCU8 E9FYK6 J9K6C7 A0A0P4Y1X2 A0A0P6F0Y2 A0A154P1E0 A0A2H8TUJ9 A0A0K8TMJ4 A0A0N8C5S3 A0A0P5D0K8 A0A0N7ZKY4 A0A224X744 A0A069DWL6 E0VK08 A0A0P5PYV0 A0A023F2M8 A0A0P4W4B1 A0A0K8VL42 A0A1A9WES6 A0A1B0FA38 B4KFT8 A0A1I8M5E1 A0A1I8PEM5 A0A0A1WFZ4 A0A0P5DRT4 A0A0P5VB79 A0A0A9XUE9 A0A0L0C426 A0A0P4YYI3 A0A3B0JYN3
PDB
3IYV
E-value=0.00615271,
Score=97
Ontologies
GO
PANTHER
Topology
Subcellular location
Endosome membrane
Late endosome
Lysosome
Golgi apparatus
trans-Golgi network
Early endosome
Cytoplasmic vesicle
Clathrin-coated vesicle
Late endosome
Lysosome
Golgi apparatus
trans-Golgi network
Early endosome
Cytoplasmic vesicle
Clathrin-coated vesicle
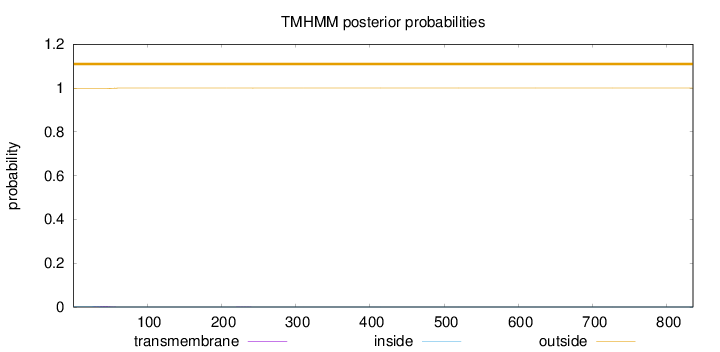
Length:
835
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0618700000000002
Exp number, first 60 AAs:
0.05539
Total prob of N-in:
0.00286
outside
1 - 835
Population Genetic Test Statistics
Pi
211.466919
Theta
170.994008
Tajima's D
0.36306
CLR
0
CSRT
0.47707614619269
Interpretation
Uncertain
