Gene
KWMTBOMO10034
Pre Gene Modal
BGIBMGA005525
Annotation
PREDICTED:_beta-1?4-galactosyltransferase_2-like_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.484
Sequence
CDS
ATGCCCAAATGTTCATTGAAAAACCGACTACTGCAACTACGGCTGAGCACTTGTGTAACAACAGTACTATGTTTAGTGGCAATTATAAATTTCTTTTCATTCTACGGAAAATACAATTACTCTCATATAAATCCACAAGACATACCCAATCAATTGTATAAAGAAATAGCTTCAAACATAGTTTTGAAAAAGTCGAAACCAGACTGTGAGTTTGAGAATATTTTGAAAAGTACAAAAACTACAGACAGCTGGGATGCGCCGAAGAAGAATGAAAACTTCAAAGCCAACAATATTGTTAACGGTAGCTATGTTCCTGAAGAATGCAACCCAATGTTCAGTGTGGCCGTGATAGTTACATACAGGAACAGACAAAAACACCTGGATGTCTTTCTACCATACATGCACAACTTTTTGAGGAAGCAACAATTGCATTATAAAATATACTTAATTGAACAAGAAGATAAAAAACCATGGAACAAAGGCTTGCTTTACAATATCGGTGCCAAAAGGGCCATAGCCGAAAAATTCCCCTGCCTCATATTGCACGATGTGGATCTTCTACCATTAGATGAAAGCAATCTCTATGCCTGCCTGAACAACCCGCGACACATGAGCGCCAGCATAGACAAGTTCAGATACGTCCTGACTTACGATTATCTAGTCGGTGGAGTGTTGGCGATAACGGCGGACCAGTTCGTGAAGGTTAATGGGTTTTCTAACAGATTCGAAGGATGGGGCGGAGAAGATGATGACTTTTACGAAAGACTGTCTGCGCACGGATTCAACGTTGTGAGGTTCCCACCTCGCATGTCGCGATATACCATGTTGGTTCACCCTCAGGAGCCAAAGAACTCGAGAAGACATCAAATTATGGCCGAAAATCTCCATACGAAAAGTCACGACGGCTACGACTCCGTGCACTACCAGACTGTAGACGAAAGACAACAGAAACTGTTCACCCAACTGCGAGTCAAGTTATGA
Protein
MPKCSLKNRLLQLRLSTCVTTVLCLVAIINFFSFYGKYNYSHINPQDIPNQLYKEIASNIVLKKSKPDCEFENILKSTKTTDSWDAPKKNENFKANNIVNGSYVPEECNPMFSVAVIVTYRNRQKHLDVFLPYMHNFLRKQQLHYKIYLIEQEDKKPWNKGLLYNIGAKRAIAEKFPCLILHDVDLLPLDESNLYACLNNPRHMSASIDKFRYVLTYDYLVGGVLAITADQFVKVNGFSNRFEGWGGEDDDFYERLSAHGFNVVRFPPRMSRYTMLVHPQEPKNSRRHQIMAENLHTKSHDGYDSVHYQTVDERQQKLFTQLRVKL
Summary
Uniprot
H9J7N3
S4PX51
A0A2A4J6Q6
A0A194QAT2
A0A0N1IQ07
A0A212FF74
+ More
A0A2H1WVA9 A0A3S2NUE5 A0A0L7LPU2 B4NF99 B4M066 A0A0A1XME8 A0A084VY14 A0A182LXD4 A0A2M3ZAW0 A0A0L0BY63 A0A182K7C5 A0A1I8PJA1 A0A182JJ51 A0A1S4GXT7 A0A182V8C6 A0A182TN22 A7UUK4 A0A1A9Z6W6 A0A1I8NAT3 A0A1A9US67 A0A182XL64 A0A182L0W7 A0A182ICG1 A0A3B0KS36 D3TQ83 W8BYL7 A0A182PCQ2 A0A1A9YBU5 A0A182W6P3 A0A336KDM0 A0A1Y9HFA8 Q0IF91 A0A182Y3F8 A0A1A9X1U6 B4KDT2 A0A182MZ00 A0A1J1HJA3 A0A0K8VID4 A0A034WD93 A0A182H5W3 A0A182QJT0 B4G4W7 Q298Z2 B3LXP0 A0A3S2N9L2 A0A1B0B842 A0A1L8E277 W5J662 A0A2M4BVI2 A0A0M4EM62 N6UAD3 B4JTB7 A0A182F4B7 A0A1W4VUD7 Q9VAQ8 B4QZF2 B3P5M7 B4PQ88 B4HZ44 D7EJY6 A0A1Y1LE70 A0A1Y1LFS1 A0A2A4JVV5 A0A2H1VH51 A0A1Y1LHY1 A0A1Y1LC51 A0A2S2QRM5 A0A1B6G8Z7 A0A1E1X7D5 A0A2R7WYP8 A0A131XG37 A0A1E1XP68 A0A1L8E1C0 A0A0L7KYX6 Q6J4T9 A0A1B6KML8 A0A1B6HT31
A0A2H1WVA9 A0A3S2NUE5 A0A0L7LPU2 B4NF99 B4M066 A0A0A1XME8 A0A084VY14 A0A182LXD4 A0A2M3ZAW0 A0A0L0BY63 A0A182K7C5 A0A1I8PJA1 A0A182JJ51 A0A1S4GXT7 A0A182V8C6 A0A182TN22 A7UUK4 A0A1A9Z6W6 A0A1I8NAT3 A0A1A9US67 A0A182XL64 A0A182L0W7 A0A182ICG1 A0A3B0KS36 D3TQ83 W8BYL7 A0A182PCQ2 A0A1A9YBU5 A0A182W6P3 A0A336KDM0 A0A1Y9HFA8 Q0IF91 A0A182Y3F8 A0A1A9X1U6 B4KDT2 A0A182MZ00 A0A1J1HJA3 A0A0K8VID4 A0A034WD93 A0A182H5W3 A0A182QJT0 B4G4W7 Q298Z2 B3LXP0 A0A3S2N9L2 A0A1B0B842 A0A1L8E277 W5J662 A0A2M4BVI2 A0A0M4EM62 N6UAD3 B4JTB7 A0A182F4B7 A0A1W4VUD7 Q9VAQ8 B4QZF2 B3P5M7 B4PQ88 B4HZ44 D7EJY6 A0A1Y1LE70 A0A1Y1LFS1 A0A2A4JVV5 A0A2H1VH51 A0A1Y1LHY1 A0A1Y1LC51 A0A2S2QRM5 A0A1B6G8Z7 A0A1E1X7D5 A0A2R7WYP8 A0A131XG37 A0A1E1XP68 A0A1L8E1C0 A0A0L7KYX6 Q6J4T9 A0A1B6KML8 A0A1B6HT31
Pubmed
19121390
23622113
26354079
22118469
26227816
17994087
+ More
25830018 24438588 26108605 12364791 25315136 20966253 20353571 24495485 17510324 25244985 25348373 26483478 15632085 20920257 23761445 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18362917 19820115 28004739 28503490 28049606 29209593 15173167
25830018 24438588 26108605 12364791 25315136 20966253 20353571 24495485 17510324 25244985 25348373 26483478 15632085 20920257 23761445 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18362917 19820115 28004739 28503490 28049606 29209593 15173167
EMBL
BABH01020434
GAIX01005648
JAA86912.1
NWSH01002778
PCG67549.1
KQ459232
+ More
KPJ02539.1 KQ460044 KPJ18304.1 AGBW02008849 OWR52382.1 ODYU01010928 SOQ56344.1 RSAL01000075 RVE48908.1 JTDY01000379 KOB77460.1 CH964251 EDW82966.1 CH940650 EDW68316.1 GBXI01001788 JAD12504.1 ATLV01018243 KE525226 KFB42858.1 AXCM01009415 GGFM01004837 MBW25588.1 JRES01001250 KNC24224.1 AAAB01008964 EDO63572.1 APCN01000798 OUUW01000013 SPP88061.1 CCAG010012178 EZ423585 ADD19861.1 GAMC01000095 JAC06461.1 UFQS01000295 UFQT01000295 SSX02567.1 SSX22941.1 CH477389 EAT41995.1 CH933806 EDW14929.1 CVRI01000001 CRK86550.1 GDHF01014029 JAI38285.1 GAKP01007229 JAC51723.1 JXUM01112743 KQ565620 KXJ70838.1 AXCN02000789 CH479179 EDW24633.1 CM000070 EAL27812.1 CH902617 EDV43934.1 RSAL01000170 RVE45184.1 JXJN01009829 GFDF01001345 JAV12739.1 ADMH02002138 ETN58350.1 GGFJ01007873 MBW57014.1 CP012526 ALC45526.1 APGK01032722 KB740848 KB632411 ENN78690.1 ERL95245.1 CH916373 EDV95007.1 AE014297 BT011457 AAF56843.1 AAR99115.1 CM000364 EDX14790.1 CH954182 EDV53277.1 CM000160 EDW98357.1 CH480819 EDW53301.1 KQ972213 EFA12917.2 GEZM01060218 GEZM01060214 JAV71188.1 GEZM01060219 GEZM01060216 JAV71190.1 NWSH01000474 PCG76155.1 ODYU01002540 SOQ40169.1 GEZM01060220 JAV71186.1 GEZM01060213 JAV71193.1 GGMS01011216 MBY80419.1 GECZ01013418 GECZ01010863 JAS56351.1 JAS58906.1 GFAC01004070 JAT95118.1 KK856012 PTY24632.1 GEFH01003511 JAP65070.1 GFAA01002312 JAU01123.1 GFDF01001551 JAV12533.1 JTDY01004223 KOB68432.1 AY601103 AAT11926.1 GEBQ01027272 JAT12705.1 GECU01029886 JAS77820.1
KPJ02539.1 KQ460044 KPJ18304.1 AGBW02008849 OWR52382.1 ODYU01010928 SOQ56344.1 RSAL01000075 RVE48908.1 JTDY01000379 KOB77460.1 CH964251 EDW82966.1 CH940650 EDW68316.1 GBXI01001788 JAD12504.1 ATLV01018243 KE525226 KFB42858.1 AXCM01009415 GGFM01004837 MBW25588.1 JRES01001250 KNC24224.1 AAAB01008964 EDO63572.1 APCN01000798 OUUW01000013 SPP88061.1 CCAG010012178 EZ423585 ADD19861.1 GAMC01000095 JAC06461.1 UFQS01000295 UFQT01000295 SSX02567.1 SSX22941.1 CH477389 EAT41995.1 CH933806 EDW14929.1 CVRI01000001 CRK86550.1 GDHF01014029 JAI38285.1 GAKP01007229 JAC51723.1 JXUM01112743 KQ565620 KXJ70838.1 AXCN02000789 CH479179 EDW24633.1 CM000070 EAL27812.1 CH902617 EDV43934.1 RSAL01000170 RVE45184.1 JXJN01009829 GFDF01001345 JAV12739.1 ADMH02002138 ETN58350.1 GGFJ01007873 MBW57014.1 CP012526 ALC45526.1 APGK01032722 KB740848 KB632411 ENN78690.1 ERL95245.1 CH916373 EDV95007.1 AE014297 BT011457 AAF56843.1 AAR99115.1 CM000364 EDX14790.1 CH954182 EDV53277.1 CM000160 EDW98357.1 CH480819 EDW53301.1 KQ972213 EFA12917.2 GEZM01060218 GEZM01060214 JAV71188.1 GEZM01060219 GEZM01060216 JAV71190.1 NWSH01000474 PCG76155.1 ODYU01002540 SOQ40169.1 GEZM01060220 JAV71186.1 GEZM01060213 JAV71193.1 GGMS01011216 MBY80419.1 GECZ01013418 GECZ01010863 JAS56351.1 JAS58906.1 GFAC01004070 JAT95118.1 KK856012 PTY24632.1 GEFH01003511 JAP65070.1 GFAA01002312 JAU01123.1 GFDF01001551 JAV12533.1 JTDY01004223 KOB68432.1 AY601103 AAT11926.1 GEBQ01027272 JAT12705.1 GECU01029886 JAS77820.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000007798 UP000008792 UP000030765 UP000075883 UP000037069 UP000075881 UP000095300 UP000075880 UP000075903 UP000075902 UP000007062 UP000092445 UP000095301 UP000078200 UP000076407 UP000075882 UP000075840 UP000268350 UP000092444 UP000075885 UP000092443 UP000075920 UP000075900 UP000008820 UP000076408 UP000091820 UP000009192 UP000075884 UP000183832 UP000069940 UP000249989 UP000075886 UP000008744 UP000001819 UP000007801 UP000092460 UP000000673 UP000092553 UP000019118 UP000030742 UP000001070 UP000069272 UP000192221 UP000000803 UP000000304 UP000008711 UP000002282 UP000001292 UP000007266
UP000037510 UP000007798 UP000008792 UP000030765 UP000075883 UP000037069 UP000075881 UP000095300 UP000075880 UP000075903 UP000075902 UP000007062 UP000092445 UP000095301 UP000078200 UP000076407 UP000075882 UP000075840 UP000268350 UP000092444 UP000075885 UP000092443 UP000075920 UP000075900 UP000008820 UP000076408 UP000091820 UP000009192 UP000075884 UP000183832 UP000069940 UP000249989 UP000075886 UP000008744 UP000001819 UP000007801 UP000092460 UP000000673 UP000092553 UP000019118 UP000030742 UP000001070 UP000069272 UP000192221 UP000000803 UP000000304 UP000008711 UP000002282 UP000001292 UP000007266
Interpro
Gene 3D
ProteinModelPortal
H9J7N3
S4PX51
A0A2A4J6Q6
A0A194QAT2
A0A0N1IQ07
A0A212FF74
+ More
A0A2H1WVA9 A0A3S2NUE5 A0A0L7LPU2 B4NF99 B4M066 A0A0A1XME8 A0A084VY14 A0A182LXD4 A0A2M3ZAW0 A0A0L0BY63 A0A182K7C5 A0A1I8PJA1 A0A182JJ51 A0A1S4GXT7 A0A182V8C6 A0A182TN22 A7UUK4 A0A1A9Z6W6 A0A1I8NAT3 A0A1A9US67 A0A182XL64 A0A182L0W7 A0A182ICG1 A0A3B0KS36 D3TQ83 W8BYL7 A0A182PCQ2 A0A1A9YBU5 A0A182W6P3 A0A336KDM0 A0A1Y9HFA8 Q0IF91 A0A182Y3F8 A0A1A9X1U6 B4KDT2 A0A182MZ00 A0A1J1HJA3 A0A0K8VID4 A0A034WD93 A0A182H5W3 A0A182QJT0 B4G4W7 Q298Z2 B3LXP0 A0A3S2N9L2 A0A1B0B842 A0A1L8E277 W5J662 A0A2M4BVI2 A0A0M4EM62 N6UAD3 B4JTB7 A0A182F4B7 A0A1W4VUD7 Q9VAQ8 B4QZF2 B3P5M7 B4PQ88 B4HZ44 D7EJY6 A0A1Y1LE70 A0A1Y1LFS1 A0A2A4JVV5 A0A2H1VH51 A0A1Y1LHY1 A0A1Y1LC51 A0A2S2QRM5 A0A1B6G8Z7 A0A1E1X7D5 A0A2R7WYP8 A0A131XG37 A0A1E1XP68 A0A1L8E1C0 A0A0L7KYX6 Q6J4T9 A0A1B6KML8 A0A1B6HT31
A0A2H1WVA9 A0A3S2NUE5 A0A0L7LPU2 B4NF99 B4M066 A0A0A1XME8 A0A084VY14 A0A182LXD4 A0A2M3ZAW0 A0A0L0BY63 A0A182K7C5 A0A1I8PJA1 A0A182JJ51 A0A1S4GXT7 A0A182V8C6 A0A182TN22 A7UUK4 A0A1A9Z6W6 A0A1I8NAT3 A0A1A9US67 A0A182XL64 A0A182L0W7 A0A182ICG1 A0A3B0KS36 D3TQ83 W8BYL7 A0A182PCQ2 A0A1A9YBU5 A0A182W6P3 A0A336KDM0 A0A1Y9HFA8 Q0IF91 A0A182Y3F8 A0A1A9X1U6 B4KDT2 A0A182MZ00 A0A1J1HJA3 A0A0K8VID4 A0A034WD93 A0A182H5W3 A0A182QJT0 B4G4W7 Q298Z2 B3LXP0 A0A3S2N9L2 A0A1B0B842 A0A1L8E277 W5J662 A0A2M4BVI2 A0A0M4EM62 N6UAD3 B4JTB7 A0A182F4B7 A0A1W4VUD7 Q9VAQ8 B4QZF2 B3P5M7 B4PQ88 B4HZ44 D7EJY6 A0A1Y1LE70 A0A1Y1LFS1 A0A2A4JVV5 A0A2H1VH51 A0A1Y1LHY1 A0A1Y1LC51 A0A2S2QRM5 A0A1B6G8Z7 A0A1E1X7D5 A0A2R7WYP8 A0A131XG37 A0A1E1XP68 A0A1L8E1C0 A0A0L7KYX6 Q6J4T9 A0A1B6KML8 A0A1B6HT31
PDB
6FWT
E-value=8.3109e-46,
Score=462
Ontologies
GO
PANTHER
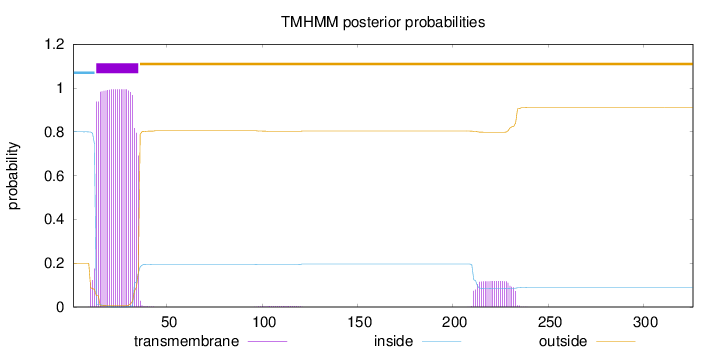
Topology
Length:
326
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.03393
Exp number, first 60 AAs:
22.45118
Total prob of N-in:
0.80010
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 326
Population Genetic Test Statistics
Pi
297.236117
Theta
197.774608
Tajima's D
1.458058
CLR
0.219889
CSRT
0.782860856957152
Interpretation
Uncertain
