Gene
KWMTBOMO10032
Pre Gene Modal
BGIBMGA005692
Annotation
ecdysone_oxidase_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.633
Sequence
CDS
ATGGTTTGCGGGTTGACTTCGTGTCTTGGGAGTGGCGCGGCCGGCGGCCTATTCTCATCCGCCGTTCAGTTCTTCGCGGCGACGCAGTGCCTGGTCGGGGAAACTTGGCCAAAAGATTCTGTGCTGCAAAATGGCTCTAGATACGACTTCATCATCGTGGGTGCCGGTACCGCTGGCTCTGCGCTCGCCGCCAGACTCTCCGAGGTTGCCAACTTCAGCGTACTCCTACTGGAGGCGGGCGGAGATCCACCGATAGAGGCTATTATACCAGCGTTCAGAGAAACTCTAAAAGCAAGTTCGGTCGATTGGAATTTTACCAGCGTGGAAAACAATATTACAAGCCAAGCCTTAAAAAGAGGAATAGAGCAACAACCTCGAGGGAAGATGCTGGGTGGAAGCGGCTCCTTGAATCACATGGTCTATGCCAGGGGTTTTCCTTCAGACTACCACGAATGGGCCTCAATCGCCGGAGAAACTTGGAACTGGACCAACGTGCTCAAATATTTCATGAAAACCGAGCACATGACGGATACGAACATTGTTAACAATCCAGAATTGATGGTGTACCACGGTCGAGGAGGTGCTATAGAAGTTTCTGGAACGAATGAAGTTATGTTCTCCATTAAGAAATTTCTACAGGCGTTCGAGGAACTTGGTTTCAAGACAGTTCCTGATATGACTTACCCAAACTCGATCGGTGCAGGATGTTTTTCGCACACCATCAGAAATGGCGAACGCGACAGCTCTCTGAGGGCTCTCCTTAACAACGCAAATAGTACAAGCCTACACATATTGAAGGATACCTTTGTAACGAAGATTATTATCGAGAACGGAACTGCAATCGGTATTGAGGCGGTAAAAGACGACAAAACGTTTTTATTTTACGCTGATCGTGAAGTCATTTTATCAGCCGGAACATTTAATACCCCGAAGTTGCTGATGCTCTCTGGCGTTGGTCGCTCAGAGCACTTGAGATCGTTAGGAATCGATGTAGTTGCAGATTTGCCGGTTGGGAGCAATCTCCACGACCACGCTATGGTATTGGCGTTTCTTGTCGCCGATAACGGGACATGCGTGTCGGATGAGGCAGAGAATTCAATGGAAGCAATAAAATATCTTTACGACAGAACTGGTTTTCTTGCCAAGGCCGACAACATGGCCGCGTATTTGCCACTCTCTAGCTCTGAGCCCACCGTACCTGAGTTTGCTCTGTACCCGACTTGTATACCACAGTTCTCGCCGTTTCGCAGCGGGTGTCTGACTTTGGGCCTGAATGAAGATTTATGCACGGAATTACACAATCTAAATCAGGAATATGAACTGGTTACAATTGCTGCTGTCCTGCTGAAACCCAAATCCAGGGGTAAAGTGGAATTGAATTCCATAAATCCATTCGACGATCCTCTAATATATGCCGGTACTTTTTCAGAAGAACAAGATCTAGATCATTTTCCTCGGTTGATCAAAATGGCTTGGTCTATTGCTGATACGAATTACTTTAGAAGCAAAAATGCACGAGTGATAAAACCTTGGGTTGAAGCTTGCAGTAATTTAACAGAGAGTGCATGGATTAAATGTATGTCAAGGGCGATGGTGACATCCGCTTGGCATTCCGTGGGTACTGCCGCTATGGGCACTGTAGTAGATGGTGACCTGAAGGTGTTAGGTATTAATGGCTTACGAGTTGTGGACGCGAGTGTAATGCCAAAAATAATTAGAGGGAACACAAATGCACCGGTTGTGATGATCGCGGAAATAGCAGCCGACCTCATTAAAGAACATTATTCTGTTAGTCGAACAGGGACAAATTTAAATAATATGACAATCGGTAATTTAACTGCCAGTAGCATGCCAAATATAAGTCAACCAAATATTAATTTAGCAGATGTTATTGAAAACAATGATATGATAAATTCTAGCCTGATAGAAGTTGAGATAACAAATGTAGAAATTATCACTACTACTGATAGGCAGTCAGACATAGATGATACTGTCAATGTCGCATGA
Protein
MVCGLTSCLGSGAAGGLFSSAVQFFAATQCLVGETWPKDSVLQNGSRYDFIIVGAGTAGSALAARLSEVANFSVLLLEAGGDPPIEAIIPAFRETLKASSVDWNFTSVENNITSQALKRGIEQQPRGKMLGGSGSLNHMVYARGFPSDYHEWASIAGETWNWTNVLKYFMKTEHMTDTNIVNNPELMVYHGRGGAIEVSGTNEVMFSIKKFLQAFEELGFKTVPDMTYPNSIGAGCFSHTIRNGERDSSLRALLNNANSTSLHILKDTFVTKIIIENGTAIGIEAVKDDKTFLFYADREVILSAGTFNTPKLLMLSGVGRSEHLRSLGIDVVADLPVGSNLHDHAMVLAFLVADNGTCVSDEAENSMEAIKYLYDRTGFLAKADNMAAYLPLSSSEPTVPEFALYPTCIPQFSPFRSGCLTLGLNEDLCTELHNLNQEYELVTIAAVLLKPKSRGKVELNSINPFDDPLIYAGTFSEEQDLDHFPRLIKMAWSIADTNYFRSKNARVIKPWVEACSNLTESAWIKCMSRAMVTSAWHSVGTAAMGTVVDGDLKVLGINGLRVVDASVMPKIIRGNTNAPVVMIAEIAADLIKEHYSVSRTGTNLNNMTIGNLTASSMPNISQPNINLADVIENNDMINSSLIEVEITNVEIITTTDRQSDIDDTVNVA
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H6AGY0
A0A088S3M1
A0A088SM03
A0A088S3S6
A0A088S3M8
A0A088S376
+ More
A0A088S3T2 A0A088SM07 A0A088S5A0 A0A0N0PE61 A0A0L7KQ89 Q95NZ0 A0A2H1VJT7 A0A1Z2RRI5 A0A194QC26 A0A2A4JGE4 A0A2W1BLG6 A0A2H1WT25 H9JYB9 A0A212FF54 A0A2A4JW72 A0A2A4JLJ2 A0A2A4JKJ5 A0A194QAU5 A0A2H1WZL6 A0A2H1VF60 A0A2H1WG27 A0A2H1VP10 A0A194QB47 A0A2W1BQ09 A0A2W1BWE7 A0A0N1IJ07 A0A2H1V9N6 A0A3S2LRK7 A0A3S2N607 A0A291P0W5 H9J861 A0A2J7PXQ0 A0A2W1BIV7 A0A2W1B5P9 A0A0L7LUK3 A0A2A4IYS9 A0A2H1V0S8 A0A212F6I0 A0A2H1WQ34 A0A2H1VF04 A0A182FZT3 A0A023EVK5 A0A182GTK3 A0A194QB82 H9JTY8 D0ABA6 A0A2H1VEJ4 A0A2A4IZE5 A0A084VW93 A0A1L8E0M0 A0A2H1VCV1 A0A1L8E037 A0A2A4IZI4 A0A2H1VDD9 A0A1B0GIG3 A0A1B0D7C5 A0A2H1VZZ7 H9JLP6 A0A2H1VF08 A0A182FZT4 A0A2K8FTP1 U5EYT3 A0A1L8E0E7 Q7QFX9 A0A1L8DZX6 A0A0L7L9T7 A0A067QT86 A0A194PSS7 A0A0L7LPB4 A0A2A4IZ69 A0A1Y9H2C4 A0A2W1B6G7 A0A1S6J0X2 A0A1B0GMI8 A0A2J7QZ44 A0A2H1VB55 A0A2H1W0I8 A0A212F947 A0A1B6EYD4 A0A194PPA0 A0A1B0DMM3 A0A2W1BPR7 H9JTY6 A0A182FZT5 W8AKH0 A0A1B6MIU6 J9JWV7 A0A0N0PDR9 A0A1B6JIE0 H9JI92 A0A2P8ZHS7 A0A2J7RBH7 A0A2A4K4K5
A0A088S3T2 A0A088SM07 A0A088S5A0 A0A0N0PE61 A0A0L7KQ89 Q95NZ0 A0A2H1VJT7 A0A1Z2RRI5 A0A194QC26 A0A2A4JGE4 A0A2W1BLG6 A0A2H1WT25 H9JYB9 A0A212FF54 A0A2A4JW72 A0A2A4JLJ2 A0A2A4JKJ5 A0A194QAU5 A0A2H1WZL6 A0A2H1VF60 A0A2H1WG27 A0A2H1VP10 A0A194QB47 A0A2W1BQ09 A0A2W1BWE7 A0A0N1IJ07 A0A2H1V9N6 A0A3S2LRK7 A0A3S2N607 A0A291P0W5 H9J861 A0A2J7PXQ0 A0A2W1BIV7 A0A2W1B5P9 A0A0L7LUK3 A0A2A4IYS9 A0A2H1V0S8 A0A212F6I0 A0A2H1WQ34 A0A2H1VF04 A0A182FZT3 A0A023EVK5 A0A182GTK3 A0A194QB82 H9JTY8 D0ABA6 A0A2H1VEJ4 A0A2A4IZE5 A0A084VW93 A0A1L8E0M0 A0A2H1VCV1 A0A1L8E037 A0A2A4IZI4 A0A2H1VDD9 A0A1B0GIG3 A0A1B0D7C5 A0A2H1VZZ7 H9JLP6 A0A2H1VF08 A0A182FZT4 A0A2K8FTP1 U5EYT3 A0A1L8E0E7 Q7QFX9 A0A1L8DZX6 A0A0L7L9T7 A0A067QT86 A0A194PSS7 A0A0L7LPB4 A0A2A4IZ69 A0A1Y9H2C4 A0A2W1B6G7 A0A1S6J0X2 A0A1B0GMI8 A0A2J7QZ44 A0A2H1VB55 A0A2H1W0I8 A0A212F947 A0A1B6EYD4 A0A194PPA0 A0A1B0DMM3 A0A2W1BPR7 H9JTY6 A0A182FZT5 W8AKH0 A0A1B6MIU6 J9JWV7 A0A0N0PDR9 A0A1B6JIE0 H9JI92 A0A2P8ZHS7 A0A2J7RBH7 A0A2A4K4K5
Pubmed
EMBL
JF433972
KF717653
KF717659
KF717662
KF717663
KF717665
+ More
KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717654 AIO02856.1 KF717652 AIO02854.1 KF717651 AIO02853.1 KF717664 AIO02866.1 KF717658 AIO02860.1 KF717661 AIO02863.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 KF717655 AIO02857.1 KQ460044 KPJ18306.1 JTDY01007102 KOB65457.1 AY035784 AY035785 AAK56551.1 AAK56552.1 ODYU01002782 SOQ40702.1 KY938814 ASA46455.1 KQ459232 KPJ02540.1 NWSH01001549 PCG70889.1 KZ150179 PZC72523.1 ODYU01010827 SOQ56167.1 BABH01042896 AGBW02008849 OWR52369.1 NWSH01000521 PCG75884.1 NWSH01001107 PCG72578.1 PCG72577.1 KPJ02554.1 ODYU01012241 SOQ58442.1 ODYU01002217 SOQ39416.1 ODYU01008368 SOQ51896.1 ODYU01003587 SOQ42538.1 KPJ02687.1 KZ150085 PZC73783.1 KZ149916 PZC77944.1 KPJ18321.1 ODYU01001405 SOQ37539.1 RSAL01000341 RVE42399.1 RVE42398.1 MF687606 ATJ44532.1 BABH01039828 NEVH01020853 PNF21103.1 KZ150015 PZC74989.1 KZ150306 PZC71499.1 JTDY01000066 KOB79079.1 NWSH01004668 PCG64789.1 ODYU01000155 SOQ34448.1 AGBW02010007 OWR49345.1 ODYU01010216 SOQ55185.1 ODYU01002218 SOQ39418.1 GAPW01000526 JAC13072.1 JXUM01087318 KQ563625 KXJ73584.1 KPJ02689.1 BABH01003799 FP102341 CBH09301.1 ODYU01001942 SOQ38842.1 NWSH01004349 PCG65195.1 ATLV01017523 KE525172 KFB42237.1 GFDF01002039 JAV12045.1 ODYU01001607 SOQ38064.1 GFDF01002040 JAV12044.1 NWSH01004383 PCG65171.1 ODYU01001944 SOQ38845.1 AJWK01014080 AJVK01026844 AJVK01026845 AJVK01026846 ODYU01005459 SOQ46343.1 BABH01005285 SOQ39415.1 KY618826 AQW43012.1 GANO01001826 JAB58045.1 GFDF01002042 JAV12042.1 AAAB01008844 EAA06000.3 GFDF01002041 JAV12043.1 JTDY01002039 KOB72263.1 KK853558 KDR06741.1 KQ459595 KPI96023.1 JTDY01000418 KOB77282.1 NWSH01004322 PCG65227.1 KZ150274 PZC71662.1 KU764422 AQS60669.1 AJVK01024561 AJVK01024562 NEVH01009076 PNF33851.1 SOQ38065.1 SOQ46342.1 AGBW02009643 OWR50262.1 GECZ01026787 JAS42982.1 KQ459603 KPI92960.1 AJVK01016891 KZ149963 PZC76281.1 BABH01003797 GAMC01020243 JAB86312.1 GEBQ01004133 JAT35844.1 ABLF02035756 ABLF02035762 KPJ18288.1 GECU01008742 JAS98964.1 BABH01022249 PYGN01000053 PSN56054.1 NEVH01005904 PNF38171.1 NWSH01000190 PCG78592.1
KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717654 AIO02856.1 KF717652 AIO02854.1 KF717651 AIO02853.1 KF717664 AIO02866.1 KF717658 AIO02860.1 KF717661 AIO02863.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 KF717655 AIO02857.1 KQ460044 KPJ18306.1 JTDY01007102 KOB65457.1 AY035784 AY035785 AAK56551.1 AAK56552.1 ODYU01002782 SOQ40702.1 KY938814 ASA46455.1 KQ459232 KPJ02540.1 NWSH01001549 PCG70889.1 KZ150179 PZC72523.1 ODYU01010827 SOQ56167.1 BABH01042896 AGBW02008849 OWR52369.1 NWSH01000521 PCG75884.1 NWSH01001107 PCG72578.1 PCG72577.1 KPJ02554.1 ODYU01012241 SOQ58442.1 ODYU01002217 SOQ39416.1 ODYU01008368 SOQ51896.1 ODYU01003587 SOQ42538.1 KPJ02687.1 KZ150085 PZC73783.1 KZ149916 PZC77944.1 KPJ18321.1 ODYU01001405 SOQ37539.1 RSAL01000341 RVE42399.1 RVE42398.1 MF687606 ATJ44532.1 BABH01039828 NEVH01020853 PNF21103.1 KZ150015 PZC74989.1 KZ150306 PZC71499.1 JTDY01000066 KOB79079.1 NWSH01004668 PCG64789.1 ODYU01000155 SOQ34448.1 AGBW02010007 OWR49345.1 ODYU01010216 SOQ55185.1 ODYU01002218 SOQ39418.1 GAPW01000526 JAC13072.1 JXUM01087318 KQ563625 KXJ73584.1 KPJ02689.1 BABH01003799 FP102341 CBH09301.1 ODYU01001942 SOQ38842.1 NWSH01004349 PCG65195.1 ATLV01017523 KE525172 KFB42237.1 GFDF01002039 JAV12045.1 ODYU01001607 SOQ38064.1 GFDF01002040 JAV12044.1 NWSH01004383 PCG65171.1 ODYU01001944 SOQ38845.1 AJWK01014080 AJVK01026844 AJVK01026845 AJVK01026846 ODYU01005459 SOQ46343.1 BABH01005285 SOQ39415.1 KY618826 AQW43012.1 GANO01001826 JAB58045.1 GFDF01002042 JAV12042.1 AAAB01008844 EAA06000.3 GFDF01002041 JAV12043.1 JTDY01002039 KOB72263.1 KK853558 KDR06741.1 KQ459595 KPI96023.1 JTDY01000418 KOB77282.1 NWSH01004322 PCG65227.1 KZ150274 PZC71662.1 KU764422 AQS60669.1 AJVK01024561 AJVK01024562 NEVH01009076 PNF33851.1 SOQ38065.1 SOQ46342.1 AGBW02009643 OWR50262.1 GECZ01026787 JAS42982.1 KQ459603 KPI92960.1 AJVK01016891 KZ149963 PZC76281.1 BABH01003797 GAMC01020243 JAB86312.1 GEBQ01004133 JAT35844.1 ABLF02035756 ABLF02035762 KPJ18288.1 GECU01008742 JAS98964.1 BABH01022249 PYGN01000053 PSN56054.1 NEVH01005904 PNF38171.1 NWSH01000190 PCG78592.1
Proteomes
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H6AGY0
A0A088S3M1
A0A088SM03
A0A088S3S6
A0A088S3M8
A0A088S376
+ More
A0A088S3T2 A0A088SM07 A0A088S5A0 A0A0N0PE61 A0A0L7KQ89 Q95NZ0 A0A2H1VJT7 A0A1Z2RRI5 A0A194QC26 A0A2A4JGE4 A0A2W1BLG6 A0A2H1WT25 H9JYB9 A0A212FF54 A0A2A4JW72 A0A2A4JLJ2 A0A2A4JKJ5 A0A194QAU5 A0A2H1WZL6 A0A2H1VF60 A0A2H1WG27 A0A2H1VP10 A0A194QB47 A0A2W1BQ09 A0A2W1BWE7 A0A0N1IJ07 A0A2H1V9N6 A0A3S2LRK7 A0A3S2N607 A0A291P0W5 H9J861 A0A2J7PXQ0 A0A2W1BIV7 A0A2W1B5P9 A0A0L7LUK3 A0A2A4IYS9 A0A2H1V0S8 A0A212F6I0 A0A2H1WQ34 A0A2H1VF04 A0A182FZT3 A0A023EVK5 A0A182GTK3 A0A194QB82 H9JTY8 D0ABA6 A0A2H1VEJ4 A0A2A4IZE5 A0A084VW93 A0A1L8E0M0 A0A2H1VCV1 A0A1L8E037 A0A2A4IZI4 A0A2H1VDD9 A0A1B0GIG3 A0A1B0D7C5 A0A2H1VZZ7 H9JLP6 A0A2H1VF08 A0A182FZT4 A0A2K8FTP1 U5EYT3 A0A1L8E0E7 Q7QFX9 A0A1L8DZX6 A0A0L7L9T7 A0A067QT86 A0A194PSS7 A0A0L7LPB4 A0A2A4IZ69 A0A1Y9H2C4 A0A2W1B6G7 A0A1S6J0X2 A0A1B0GMI8 A0A2J7QZ44 A0A2H1VB55 A0A2H1W0I8 A0A212F947 A0A1B6EYD4 A0A194PPA0 A0A1B0DMM3 A0A2W1BPR7 H9JTY6 A0A182FZT5 W8AKH0 A0A1B6MIU6 J9JWV7 A0A0N0PDR9 A0A1B6JIE0 H9JI92 A0A2P8ZHS7 A0A2J7RBH7 A0A2A4K4K5
A0A088S3T2 A0A088SM07 A0A088S5A0 A0A0N0PE61 A0A0L7KQ89 Q95NZ0 A0A2H1VJT7 A0A1Z2RRI5 A0A194QC26 A0A2A4JGE4 A0A2W1BLG6 A0A2H1WT25 H9JYB9 A0A212FF54 A0A2A4JW72 A0A2A4JLJ2 A0A2A4JKJ5 A0A194QAU5 A0A2H1WZL6 A0A2H1VF60 A0A2H1WG27 A0A2H1VP10 A0A194QB47 A0A2W1BQ09 A0A2W1BWE7 A0A0N1IJ07 A0A2H1V9N6 A0A3S2LRK7 A0A3S2N607 A0A291P0W5 H9J861 A0A2J7PXQ0 A0A2W1BIV7 A0A2W1B5P9 A0A0L7LUK3 A0A2A4IYS9 A0A2H1V0S8 A0A212F6I0 A0A2H1WQ34 A0A2H1VF04 A0A182FZT3 A0A023EVK5 A0A182GTK3 A0A194QB82 H9JTY8 D0ABA6 A0A2H1VEJ4 A0A2A4IZE5 A0A084VW93 A0A1L8E0M0 A0A2H1VCV1 A0A1L8E037 A0A2A4IZI4 A0A2H1VDD9 A0A1B0GIG3 A0A1B0D7C5 A0A2H1VZZ7 H9JLP6 A0A2H1VF08 A0A182FZT4 A0A2K8FTP1 U5EYT3 A0A1L8E0E7 Q7QFX9 A0A1L8DZX6 A0A0L7L9T7 A0A067QT86 A0A194PSS7 A0A0L7LPB4 A0A2A4IZ69 A0A1Y9H2C4 A0A2W1B6G7 A0A1S6J0X2 A0A1B0GMI8 A0A2J7QZ44 A0A2H1VB55 A0A2H1W0I8 A0A212F947 A0A1B6EYD4 A0A194PPA0 A0A1B0DMM3 A0A2W1BPR7 H9JTY6 A0A182FZT5 W8AKH0 A0A1B6MIU6 J9JWV7 A0A0N0PDR9 A0A1B6JIE0 H9JI92 A0A2P8ZHS7 A0A2J7RBH7 A0A2A4K4K5
PDB
5OC1
E-value=4.81437e-57,
Score=562
Ontologies
GO
Topology
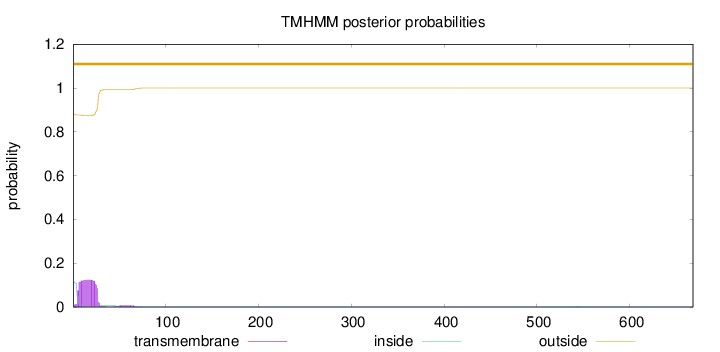
Length:
668
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.87558
Exp number, first 60 AAs:
2.81947
Total prob of N-in:
0.12210
outside
1 - 668
Population Genetic Test Statistics
Pi
232.224024
Theta
199.854972
Tajima's D
0.500731
CLR
0.339236
CSRT
0.516674166291685
Interpretation
Uncertain
