Pre Gene Modal
BGIBMGA005702
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup85_[Amyelois_transitella]
Full name
Nuclear pore complex protein Nup85
+ More
Nuclear pore complex protein Nup75
Nuclear pore complex protein Nup75
Alternative Name
Nucleoporin Nup85
Location in the cell
Mitochondrial Reliability : 1.243 PlasmaMembrane Reliability : 1.343
Sequence
CDS
ATGAATATAACGACTTTTTCTAACAATAATGTTCGTGGCGATGATAGTAGTCCTATTATTCGAGCTAAAACATTTAATATACCAGAAAAATGTTTCAATAAGGGCGTAGGATGTGCGTGGAGGAGTGGTAACAAGTTTTCTATCTACCCACGCAGTCAAACTGTTTCGAAAACACCAAAAAATGATGTCGAAAACAAAATACTCAATATACGTCACGAGATTGTATTATTTCATCCGATTTTACGAAAACTGGTAAACGAATCCAATGGTACATATTTATCTCTGCAGAAACTCGTCGATTCTTCAAAAAATAATGACAATCACGTGGAATTATTAAAACTTTCACGACAATATCGTTCTATTATAAGAGTGTGCATTGAAAACTTGCAAGAAGCCCTTAATGATGCGAAAGACGGCAGCGACATGATCAATTTACAATCATACATTACAATATTTTATTCTGTTGAATGTATTTGGCATTTATGTGAGATCCTTTACATAGATAAGATTCCCGGAGACATTGTGTTGCCATACCTCCTAGAATGGGTTCGATTCCATTTTCCGTGCCATGAGCAGACTGCAGCGCAGTTGCTGGAGGCCTGTGAGCGAGGTTCAGAAGACCACGATGACTATTGGGATACTGTGATTGGCATGGTTGTGCAAGGGCGAGTAGATGTCGCCCGAGCTCTACTGAAATTAAATTCATTATCAGAAACAAATGAATTTAAGCTTGTTGATAATTCATTACGGAGTATGCCTGTTTATAGTATATATGGTGGTATATCAACTGGGGAGTTTACAGTAACTTGGAAGCATTGGCAGGCTGAATGCAGAGCTAAGGTTACTAGCAGAGTGTTGGCATCACAACCAAAACTTGAACTTATTATGAGATTAATCATAGGAGAATATTCAGCATTTGAGTCAATACGTAGCACATACACATCGTGGTTTGATATGTTGGGAGGCTACGTAATGTTCACAGCACCTTGGTCTAGGACTCATGAGTTGTCTGCTGCAGCATCGGCCTGTGCTGGAATGGGATCTATGCCCTCAGGCTCTAGGCTGGATAGGACTTTACGGGCATTGTTGGAAGGTGATATGCATCAAGTAATTCATGAAATCCAGCATATAACAGATAATGGATGGTTTGCAACACATCTATCAGACATATTGTATCATTGTGGAAAATTAAAAGTATTGGATAAACACCAAAATAATGTAACTCAACGTCTCCGAGACAGTTTAATTCTTGAATATGGAACCTTGATAATGGAGCACAAGTCACTCTGGTCCGTCGGACTATCCTACCTTGCTTCATGTCCCCCTGAAGGTCTATGCCGAGCTCATTTGTTGCTAGAGAGGATTCCGATCAATACTCATGCCAAAGCTATGAGGGTTGTTGCTGAAGCTAATAAATATGGACTTTTGGAAGTTGGCGAGGGCGTGTGGCGCGTGTTGTCGGGGCGGCACAGCGCGGCGGGGCGGGACGGCGCGGCGCTGAGCGCGGCGGTGCGCGGCGGCGCGGCGGGGCGGGCGGGGCGCTGCGCCGGGGCCGCGCTGCGCCACTACTGCCGCACCGGCTGCCTGCCCGCGCCGGACCTGCTGCTCACCGCCGGACCGGCGCTGCTCATTGACGACACTCTACTCTTCTTAGGTAAATATTGTGAATTTCACCGCATGTACAAGAATAAAGAGTTCCGAAAAGCCGCACAGCTCCTCATATCGCTCATCACATCTAAAATAGCCCCGGATTTTTTCTGGGAAACCTTACTGCTAGATGCTCTACCGCTATTGGAATCTGACGAGGCCATGTTCACGGCAGATGACACATACGATATGATGCTGTGCCTTGAGCTACGAGCCCAGAACTTCAACCGAGAGAAAGCGGAACTCCTCAGATTGGCTTTACTTAGGAACCTGGCACGGACTGCACTAGCCGAGAAACCTGAAGAAAGTACTCAGTGA
Protein
MNITTFSNNNVRGDDSSPIIRAKTFNIPEKCFNKGVGCAWRSGNKFSIYPRSQTVSKTPKNDVENKILNIRHEIVLFHPILRKLVNESNGTYLSLQKLVDSSKNNDNHVELLKLSRQYRSIIRVCIENLQEALNDAKDGSDMINLQSYITIFYSVECIWHLCEILYIDKIPGDIVLPYLLEWVRFHFPCHEQTAAQLLEACERGSEDHDDYWDTVIGMVVQGRVDVARALLKLNSLSETNEFKLVDNSLRSMPVYSIYGGISTGEFTVTWKHWQAECRAKVTSRVLASQPKLELIMRLIIGEYSAFESIRSTYTSWFDMLGGYVMFTAPWSRTHELSAAASACAGMGSMPSGSRLDRTLRALLEGDMHQVIHEIQHITDNGWFATHLSDILYHCGKLKVLDKHQNNVTQRLRDSLILEYGTLIMEHKSLWSVGLSYLASCPPEGLCRAHLLLERIPINTHAKAMRVVAEANKYGLLEVGEGVWRVLSGRHSAAGRDGAALSAAVRGGAAGRAGRCAGAALRHYCRTGCLPAPDLLLTAGPALLIDDTLLFLGKYCEFHRMYKNKEFRKAAQLLISLITSKIAPDFFWETLLLDALPLLESDEAMFTADDTYDMMLCLELRAQNFNREKAELLRLALLRNLARTALAEKPEESTQ
Summary
Description
Functions as a component of the nuclear pore complex (NPC).
Subunit
Component of the nuclear pore complex (NPC).
Similarity
Belongs to the nucleoporin Nup85 family.
Uniprot
H9J860
A0A2A4JTY9
A0A2W1BDW9
A0A212FF43
A0A0N0PDV8
A0A194QAU0
+ More
D6WUN5 A0A1W4WWR0 J3JZJ7 A0A0C9RRP8 A0A232F2F9 K7IZ68 A0A1Y1LCD5 U4UCW5 A0A2J7QAI3 E9ILL6 E2BQ39 A0A067RBY6 E2A230 A0A195F910 F4WFC2 A0A158NCN6 A0A195CG66 A0A151XF36 A0A0L7R102 A0A3L8D3Q4 A0A0T6B2G1 A0A088A8J6 A0A2A3E3D4 A0A026VXJ3 A0A0M9A590 A0A1I8NLQ6 A0A1B6GHG6 A0A0K8TWZ2 W8BWM4 A0A1I8MCV3 A0A034VNQ2 A0A0A1X0B0 A0A1B6DZI9 A0A1L8DXE2 E0VCB4 B0WI27 Q16KI2 A0A1Q3F1J2 A0A182HB70 A0A2P8YJZ7 A0A1J1I3J6 A0A1B6L7M7 A0A1A9VQW1 A0A1A9W0U1 A0A195BYM2 A0A1A9Z0L4 A0A195EM98 A0A1B0GBI6 A0A1B0BYI5 A0A0P5XKQ7 A0A0N8CSH8 A0A0P5KET1 A0A0N8B669 A0A0P5V7M5 A0A0N8AG70 A0A0P4XYD5 A0A0P6GML0 A0A336M568 A0A0N8BLU2 A0A0N8E845 A0A0P6E8K6 A0A336MU20 B4LKX5 A0A3Q3GR61 A0A3Q3GTH7 A0A0P5DIJ6 A0A0P5TYW5 A0A0P6A7Z2 A0A0P5UTT0 A0A0P5WQQ6 A0A0P5BVZ7 A0A0P5STU0 A0A0P5C3T7 A0A0P5VDT2 A0A0P5X226 A0A0P5I5D8 A0A0P6H4Z2 A0A0P6IR56 A0A0P6G969 A0A3B4TBL9 W5N750 A0A0P5WG31 A0A0P5ZG10 A0A0P5B5Y7 W5N743 A0A2U9CQF3 A0A3Q1JWV5 A0A0P5AEH7 A0A0P5WLG6 A0A0P5YZE4
D6WUN5 A0A1W4WWR0 J3JZJ7 A0A0C9RRP8 A0A232F2F9 K7IZ68 A0A1Y1LCD5 U4UCW5 A0A2J7QAI3 E9ILL6 E2BQ39 A0A067RBY6 E2A230 A0A195F910 F4WFC2 A0A158NCN6 A0A195CG66 A0A151XF36 A0A0L7R102 A0A3L8D3Q4 A0A0T6B2G1 A0A088A8J6 A0A2A3E3D4 A0A026VXJ3 A0A0M9A590 A0A1I8NLQ6 A0A1B6GHG6 A0A0K8TWZ2 W8BWM4 A0A1I8MCV3 A0A034VNQ2 A0A0A1X0B0 A0A1B6DZI9 A0A1L8DXE2 E0VCB4 B0WI27 Q16KI2 A0A1Q3F1J2 A0A182HB70 A0A2P8YJZ7 A0A1J1I3J6 A0A1B6L7M7 A0A1A9VQW1 A0A1A9W0U1 A0A195BYM2 A0A1A9Z0L4 A0A195EM98 A0A1B0GBI6 A0A1B0BYI5 A0A0P5XKQ7 A0A0N8CSH8 A0A0P5KET1 A0A0N8B669 A0A0P5V7M5 A0A0N8AG70 A0A0P4XYD5 A0A0P6GML0 A0A336M568 A0A0N8BLU2 A0A0N8E845 A0A0P6E8K6 A0A336MU20 B4LKX5 A0A3Q3GR61 A0A3Q3GTH7 A0A0P5DIJ6 A0A0P5TYW5 A0A0P6A7Z2 A0A0P5UTT0 A0A0P5WQQ6 A0A0P5BVZ7 A0A0P5STU0 A0A0P5C3T7 A0A0P5VDT2 A0A0P5X226 A0A0P5I5D8 A0A0P6H4Z2 A0A0P6IR56 A0A0P6G969 A0A3B4TBL9 W5N750 A0A0P5WG31 A0A0P5ZG10 A0A0P5B5Y7 W5N743 A0A2U9CQF3 A0A3Q1JWV5 A0A0P5AEH7 A0A0P5WLG6 A0A0P5YZE4
Pubmed
EMBL
BABH01039826
BABH01039827
NWSH01000621
PCG75246.1
KZ150306
PZC71497.1
+ More
AGBW02008849 OWR52371.1 KQ460044 KPJ18315.1 KQ459232 KPJ02549.1 KQ971363 EFA07823.1 BT128679 AEE63636.1 GBYB01010046 JAG79813.1 NNAY01001222 OXU24689.1 GEZM01061334 GEZM01061333 JAV70558.1 KB632061 ERL88426.1 NEVH01016330 PNF25587.1 GL764129 EFZ18439.1 GL449694 EFN82227.1 KK852804 KDR16208.1 GL435917 EFN72514.1 KQ981727 KYN36928.1 GL888115 EGI67173.1 ADTU01012001 KQ977791 KYM99712.1 KQ982215 KYQ58945.1 KQ414669 KOC64476.1 QOIP01000014 RLU15030.1 LJIG01016187 KRT81355.1 KZ288419 PBC25994.1 KK107609 EZA48492.1 KQ435732 KOX77505.1 GECZ01007890 JAS61879.1 GDHF01033310 JAI19004.1 GAMC01012960 GAMC01012958 GAMC01012955 GAMC01012951 JAB93604.1 GAKP01015779 JAC43173.1 GBXI01012657 GBXI01009735 JAD01635.1 JAD04557.1 GEDC01006224 JAS31074.1 GFDF01002966 JAV11118.1 DS235051 EEB11000.1 DS231941 EDS28109.1 CH477957 EAT34798.1 GFDL01013619 JAV21426.1 JXUM01031797 JXUM01031798 JXUM01031799 KQ560936 KXJ80270.1 PYGN01000541 PSN44565.1 CVRI01000040 CRK94771.1 GEBQ01020280 JAT19697.1 KQ976396 KYM93033.1 KQ978691 KYN29251.1 CCAG010014016 JXJN01022668 GDIP01082953 JAM20762.1 GDIP01103223 GDIP01053401 LRGB01001411 JAM00492.1 KZS11984.1 GDIQ01210951 JAK40774.1 GDIQ01208907 JAK42818.1 GDIP01105930 JAL97784.1 GDIP01150577 JAJ72825.1 GDIP01235134 JAI88267.1 GDIQ01031477 JAN63260.1 UFQT01000574 SSX25414.1 GDIQ01165123 JAK86602.1 GDIQ01052508 JAN42229.1 GDIQ01072191 JAN22546.1 UFQT01002523 SSX33620.1 CH940648 EDW60779.1 GDIP01155693 JAJ67709.1 GDIP01120589 JAL83125.1 GDIP01034345 JAM69370.1 GDIP01108428 JAL95286.1 GDIP01082952 JAM20763.1 GDIP01183291 JAJ40111.1 GDIP01169080 GDIP01140642 JAL63072.1 GDIP01191584 JAJ31818.1 GDIP01103224 JAM00491.1 GDIP01078450 JAM25265.1 GDIQ01220000 JAK31725.1 GDIQ01024554 JAN70183.1 GDIQ01002830 JAN91907.1 GDIQ01039511 JAN55226.1 AHAT01000429 GDIP01087289 JAM16426.1 GDIP01044595 JAM59120.1 GDIP01193582 GDIP01060999 JAJ29820.1 CP026261 AWP18828.1 GDIP01200884 JAJ22518.1 GDIP01084748 JAM18967.1 GDIP01051004 JAM52711.1
AGBW02008849 OWR52371.1 KQ460044 KPJ18315.1 KQ459232 KPJ02549.1 KQ971363 EFA07823.1 BT128679 AEE63636.1 GBYB01010046 JAG79813.1 NNAY01001222 OXU24689.1 GEZM01061334 GEZM01061333 JAV70558.1 KB632061 ERL88426.1 NEVH01016330 PNF25587.1 GL764129 EFZ18439.1 GL449694 EFN82227.1 KK852804 KDR16208.1 GL435917 EFN72514.1 KQ981727 KYN36928.1 GL888115 EGI67173.1 ADTU01012001 KQ977791 KYM99712.1 KQ982215 KYQ58945.1 KQ414669 KOC64476.1 QOIP01000014 RLU15030.1 LJIG01016187 KRT81355.1 KZ288419 PBC25994.1 KK107609 EZA48492.1 KQ435732 KOX77505.1 GECZ01007890 JAS61879.1 GDHF01033310 JAI19004.1 GAMC01012960 GAMC01012958 GAMC01012955 GAMC01012951 JAB93604.1 GAKP01015779 JAC43173.1 GBXI01012657 GBXI01009735 JAD01635.1 JAD04557.1 GEDC01006224 JAS31074.1 GFDF01002966 JAV11118.1 DS235051 EEB11000.1 DS231941 EDS28109.1 CH477957 EAT34798.1 GFDL01013619 JAV21426.1 JXUM01031797 JXUM01031798 JXUM01031799 KQ560936 KXJ80270.1 PYGN01000541 PSN44565.1 CVRI01000040 CRK94771.1 GEBQ01020280 JAT19697.1 KQ976396 KYM93033.1 KQ978691 KYN29251.1 CCAG010014016 JXJN01022668 GDIP01082953 JAM20762.1 GDIP01103223 GDIP01053401 LRGB01001411 JAM00492.1 KZS11984.1 GDIQ01210951 JAK40774.1 GDIQ01208907 JAK42818.1 GDIP01105930 JAL97784.1 GDIP01150577 JAJ72825.1 GDIP01235134 JAI88267.1 GDIQ01031477 JAN63260.1 UFQT01000574 SSX25414.1 GDIQ01165123 JAK86602.1 GDIQ01052508 JAN42229.1 GDIQ01072191 JAN22546.1 UFQT01002523 SSX33620.1 CH940648 EDW60779.1 GDIP01155693 JAJ67709.1 GDIP01120589 JAL83125.1 GDIP01034345 JAM69370.1 GDIP01108428 JAL95286.1 GDIP01082952 JAM20763.1 GDIP01183291 JAJ40111.1 GDIP01169080 GDIP01140642 JAL63072.1 GDIP01191584 JAJ31818.1 GDIP01103224 JAM00491.1 GDIP01078450 JAM25265.1 GDIQ01220000 JAK31725.1 GDIQ01024554 JAN70183.1 GDIQ01002830 JAN91907.1 GDIQ01039511 JAN55226.1 AHAT01000429 GDIP01087289 JAM16426.1 GDIP01044595 JAM59120.1 GDIP01193582 GDIP01060999 JAJ29820.1 CP026261 AWP18828.1 GDIP01200884 JAJ22518.1 GDIP01084748 JAM18967.1 GDIP01051004 JAM52711.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000192223 UP000215335 UP000002358 UP000030742 UP000235965 UP000008237 UP000027135 UP000000311 UP000078541 UP000007755 UP000005205 UP000078542 UP000075809 UP000053825 UP000279307 UP000005203 UP000242457 UP000053097 UP000053105 UP000095300 UP000095301 UP000009046 UP000002320 UP000008820 UP000069940 UP000249989 UP000245037 UP000183832 UP000078200 UP000091820 UP000078540 UP000092445 UP000078492 UP000092444 UP000092460 UP000076858 UP000008792 UP000261660 UP000261420 UP000018468 UP000246464 UP000265040
UP000192223 UP000215335 UP000002358 UP000030742 UP000235965 UP000008237 UP000027135 UP000000311 UP000078541 UP000007755 UP000005205 UP000078542 UP000075809 UP000053825 UP000279307 UP000005203 UP000242457 UP000053097 UP000053105 UP000095300 UP000095301 UP000009046 UP000002320 UP000008820 UP000069940 UP000249989 UP000245037 UP000183832 UP000078200 UP000091820 UP000078540 UP000092445 UP000078492 UP000092444 UP000092460 UP000076858 UP000008792 UP000261660 UP000261420 UP000018468 UP000246464 UP000265040
PRIDE
Pfam
PF07575 Nucleopor_Nup85
Interpro
IPR011502
Nucleoporin_Nup85
ProteinModelPortal
H9J860
A0A2A4JTY9
A0A2W1BDW9
A0A212FF43
A0A0N0PDV8
A0A194QAU0
+ More
D6WUN5 A0A1W4WWR0 J3JZJ7 A0A0C9RRP8 A0A232F2F9 K7IZ68 A0A1Y1LCD5 U4UCW5 A0A2J7QAI3 E9ILL6 E2BQ39 A0A067RBY6 E2A230 A0A195F910 F4WFC2 A0A158NCN6 A0A195CG66 A0A151XF36 A0A0L7R102 A0A3L8D3Q4 A0A0T6B2G1 A0A088A8J6 A0A2A3E3D4 A0A026VXJ3 A0A0M9A590 A0A1I8NLQ6 A0A1B6GHG6 A0A0K8TWZ2 W8BWM4 A0A1I8MCV3 A0A034VNQ2 A0A0A1X0B0 A0A1B6DZI9 A0A1L8DXE2 E0VCB4 B0WI27 Q16KI2 A0A1Q3F1J2 A0A182HB70 A0A2P8YJZ7 A0A1J1I3J6 A0A1B6L7M7 A0A1A9VQW1 A0A1A9W0U1 A0A195BYM2 A0A1A9Z0L4 A0A195EM98 A0A1B0GBI6 A0A1B0BYI5 A0A0P5XKQ7 A0A0N8CSH8 A0A0P5KET1 A0A0N8B669 A0A0P5V7M5 A0A0N8AG70 A0A0P4XYD5 A0A0P6GML0 A0A336M568 A0A0N8BLU2 A0A0N8E845 A0A0P6E8K6 A0A336MU20 B4LKX5 A0A3Q3GR61 A0A3Q3GTH7 A0A0P5DIJ6 A0A0P5TYW5 A0A0P6A7Z2 A0A0P5UTT0 A0A0P5WQQ6 A0A0P5BVZ7 A0A0P5STU0 A0A0P5C3T7 A0A0P5VDT2 A0A0P5X226 A0A0P5I5D8 A0A0P6H4Z2 A0A0P6IR56 A0A0P6G969 A0A3B4TBL9 W5N750 A0A0P5WG31 A0A0P5ZG10 A0A0P5B5Y7 W5N743 A0A2U9CQF3 A0A3Q1JWV5 A0A0P5AEH7 A0A0P5WLG6 A0A0P5YZE4
D6WUN5 A0A1W4WWR0 J3JZJ7 A0A0C9RRP8 A0A232F2F9 K7IZ68 A0A1Y1LCD5 U4UCW5 A0A2J7QAI3 E9ILL6 E2BQ39 A0A067RBY6 E2A230 A0A195F910 F4WFC2 A0A158NCN6 A0A195CG66 A0A151XF36 A0A0L7R102 A0A3L8D3Q4 A0A0T6B2G1 A0A088A8J6 A0A2A3E3D4 A0A026VXJ3 A0A0M9A590 A0A1I8NLQ6 A0A1B6GHG6 A0A0K8TWZ2 W8BWM4 A0A1I8MCV3 A0A034VNQ2 A0A0A1X0B0 A0A1B6DZI9 A0A1L8DXE2 E0VCB4 B0WI27 Q16KI2 A0A1Q3F1J2 A0A182HB70 A0A2P8YJZ7 A0A1J1I3J6 A0A1B6L7M7 A0A1A9VQW1 A0A1A9W0U1 A0A195BYM2 A0A1A9Z0L4 A0A195EM98 A0A1B0GBI6 A0A1B0BYI5 A0A0P5XKQ7 A0A0N8CSH8 A0A0P5KET1 A0A0N8B669 A0A0P5V7M5 A0A0N8AG70 A0A0P4XYD5 A0A0P6GML0 A0A336M568 A0A0N8BLU2 A0A0N8E845 A0A0P6E8K6 A0A336MU20 B4LKX5 A0A3Q3GR61 A0A3Q3GTH7 A0A0P5DIJ6 A0A0P5TYW5 A0A0P6A7Z2 A0A0P5UTT0 A0A0P5WQQ6 A0A0P5BVZ7 A0A0P5STU0 A0A0P5C3T7 A0A0P5VDT2 A0A0P5X226 A0A0P5I5D8 A0A0P6H4Z2 A0A0P6IR56 A0A0P6G969 A0A3B4TBL9 W5N750 A0A0P5WG31 A0A0P5ZG10 A0A0P5B5Y7 W5N743 A0A2U9CQF3 A0A3Q1JWV5 A0A0P5AEH7 A0A0P5WLG6 A0A0P5YZE4
PDB
5A9Q
E-value=8.6976e-87,
Score=819
Ontologies
KEGG
GO
PANTHER
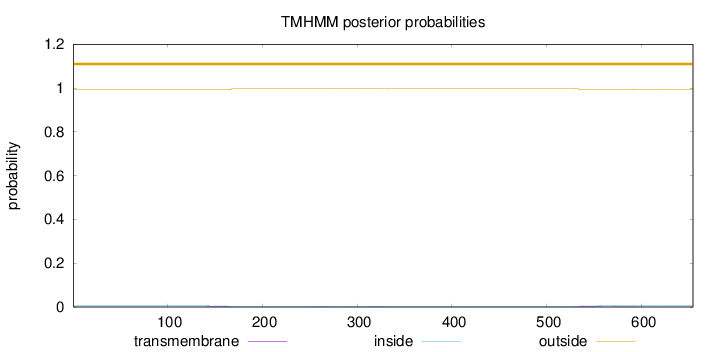
Topology
Subcellular location
Length:
654
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26318
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00668
outside
1 - 654
Population Genetic Test Statistics
Pi
228.091546
Theta
191.51148
Tajima's D
2.179472
CLR
0.34447
CSRT
0.909204539773011
Interpretation
Uncertain
