Gene
KWMTBOMO10020
Pre Gene Modal
BGIBMGA005699
Annotation
PREDICTED:_7?8-dihydro-8-oxoguanine_triphosphatase-like_[Papilio_polytes]
Full name
7,8-dihydro-8-oxoguanine triphosphatase
Alternative Name
2-hydroxy-dATP diphosphatase
8-oxo-dGTPase
Nucleoside diphosphate-linked moiety X motif 1
8-oxo-dGTPase
Nucleoside diphosphate-linked moiety X motif 1
Location in the cell
Cytoplasmic Reliability : 2.469
Sequence
CDS
ATGCTTCTAAAGAAAGTATACACTCTAGTCTTCATCTCGAAAGACCAACAAATACTTTTGGGTCTTAAAAAGCGCGGGTTTGGCGTCAACAAATGGAATGGTTTTGGCGGTAAATTGGAGCCGAACGAAACAATAGTTGAGGCCGCTTCAAGAGAATTACAAGAAGAATGCTGTATTAGGGTAAAAACGACAGATTTAAAGAACATCGGACATTTAGAATTTACTTTTGAAGGGGATAAAACTATGATGGAAGTCAGAGTATTTGGTTCGAAATTGTTTGAAGGAATTCCTAGAGAGACCGAAGAAATGTTACCGAAATGGTTTCCTTGCAATAACATCCCTTTTGATAACATGTGGCCTGATGATAGAATTTGGTTTCCCTATATGCTGAAGGGACAACTGTTTTATGGTAGATTTCATTACAAAGATCTTAATACAATATTGAATTATCATATTGAAGAGCTGAAGTCTATGGAAGAATTTTATAATAGAAATTTGTAA
Protein
MLLKKVYTLVFISKDQQILLGLKKRGFGVNKWNGFGGKLEPNETIVEAASRELQEECCIRVKTTDLKNIGHLEFTFEGDKTMMEVRVFGSKLFEGIPRETEEMLPKWFPCNNIPFDNMWPDDRIWFPYMLKGQLFYGRFHYKDLNTILNYHIEELKSMEEFYNRNL
Summary
Description
Antimutagenic (PubMed:11572992). Plays a redundant role in sanitizing oxidized nucleotide pools, such as 8-oxo-dGTP pools (PubMed:29281266). Acts as a sanitizing enzyme for oxidized nucleotide pools, thus suppressing cell dysfunction and death induced by oxidative stress. Hydrolyzes 8-oxo-dGTP, 8-oxo-dATP and 2-OH-dATP, thus preventing misincorporation of oxidized purine nucleoside triphosphates into DNA and subsequently preventing A:T to C:G and G:C to T:A transversions. Able to hydrolyze also the corresponding ribonucleotides, 2-OH-ATP, 8-oxo-GTP and 8-oxo-ATP (By similarity). Does not play a role in U8 snoRNA decapping activity (PubMed:21070968). Binds U8 snoRNA (PubMed:21070968).
Antimutagenic. Plays a redundant role in sanitizing oxidized nucleotide pools, such as 8-oxo-dGTP pools. Acts as a sanitizing enzyme for oxidized nucleotide pools, thus suppressing cell dysfunction and death induced by oxidative stress (PubMed:7586133). Hydrolyzes 8-oxo-dGTP, 8-oxo-dATP and 2-OH-dATP, thus preventing misincorporation of oxidized purine nucleoside triphosphates into DNA and subsequently preventing A:T to C:G and G:C to T:A transversions. Able to hydrolyze also the corresponding ribonucleotides, 2-OH-ATP, 8-oxo-GTP and 8-oxo-ATP (By similarity). Does not play a role in U8 snoRNA decapping activity. Binds U8 snoRNA (By similarity).
Antimutagenic. Plays a redundant role in sanitizing oxidized nucleotide pools, such as 8-oxo-dGTP pools. Acts as a sanitizing enzyme for oxidized nucleotide pools, thus suppressing cell dysfunction and death induced by oxidative stress (PubMed:7586133). Hydrolyzes 8-oxo-dGTP, 8-oxo-dATP and 2-OH-dATP, thus preventing misincorporation of oxidized purine nucleoside triphosphates into DNA and subsequently preventing A:T to C:G and G:C to T:A transversions. Able to hydrolyze also the corresponding ribonucleotides, 2-OH-ATP, 8-oxo-GTP and 8-oxo-ATP (By similarity). Does not play a role in U8 snoRNA decapping activity. Binds U8 snoRNA (By similarity).
Catalytic Activity
8-oxo-dGTP + H2O = 8-oxo-dGMP + diphosphate + H(+)
2-hydroxy-dATP + H2O = 2-hydroxy-dAMP + diphosphate + H(+)
2-hydroxy-dATP + H2O = 2-hydroxy-dAMP + diphosphate + H(+)
Cofactor
Mg(2+)
Subunit
Monomer.
Similarity
Belongs to the Nudix hydrolase family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Cytoplasmic vesicle
Hydrolase
Magnesium
Membrane
Metal-binding
Nucleus
Reference proteome
RNA-binding
Signal
Feature
chain 7,8-dihydro-8-oxoguanine triphosphatase
Uniprot
H9J857
A0A0N0PDS7
A0A2H1X285
A0A212FF71
A0A2W1B7M3
A0A2A4JT84
+ More
A0A0L7LI40 A0A194QAQ3 S4PB70 C3ZJU9 W4Z1F4 A0A0P4WJF1 A0A2P2I8N0 A0A2B4SW84 A0A0B7B346 A0A232F278 A0A088A8N2 E9GZ80 A0A2G8JUD6 K7IZ70 G3WX39 S7QBG4 I3MPS7 A0A0M9A593 H9GEQ8 A0A2T7PZD0 F7FR03 A0A1W4Y9D1 A0A0P6AKK3 L5LRE6 A0A077WVB4 A0A210QSX7 A0A315V3X5 A0A0P5N7B8 G5BI84 H0VRB9 V9LD01 A0A1J4WNZ2 A0A2Y9GDT1 K9IQL2 A0A0L7R0U7 A0A091DB69 K1PBH8 A0A2L2Y921 A0A3B5MEY7 W5LZU0 A0A3B3SEY4 A0A0G0U9Y9 A0A0F8C880 L8Y3L9 A0A3B4X7C0 A0A2J7QAH4 P53368 A0A3L7HQ12 A0A3B4V3R6 A0A2Y9J9K4 A0A250YAK2 A0A226EFD6 A0A340Y2C8 A0A3B3B4G3 U6D127 A0A3Q4BXF9 A0A3Q7NCN7 A0A2U3VHB8 M3Z2J0 P53369 A0A2M7RHS1 T1J6Y5 A0A3Q1B3G7 A0A3P8THN9 A0A1S3JEB6 A0A2P4X9Z8 A0A2U9CT94 G1LAD2 F7IL36 A0A287AR30 A0A2U3XF28 H3CJ26 A0A1F7U7E9 A0A1D2MTM7 A0A3P8X2A5 W5P5K1 A0A2K6USY0 A0A067R9D0 A0A146XTG5 A0A1A6GSK5 A0A3B3YP22 A0A087UUR3 A0A146NAI6 A0A341CX57 A0A3P9JNP2 A0A024UP77 A0A2U4CBW9 A0A2Y9MQ33 A0A1U7T6Y9 A0A3B3I653 A0A384CNV9 A0A3Q3FRG5 A0A087YAZ4 A0A3B3VKL9
A0A0L7LI40 A0A194QAQ3 S4PB70 C3ZJU9 W4Z1F4 A0A0P4WJF1 A0A2P2I8N0 A0A2B4SW84 A0A0B7B346 A0A232F278 A0A088A8N2 E9GZ80 A0A2G8JUD6 K7IZ70 G3WX39 S7QBG4 I3MPS7 A0A0M9A593 H9GEQ8 A0A2T7PZD0 F7FR03 A0A1W4Y9D1 A0A0P6AKK3 L5LRE6 A0A077WVB4 A0A210QSX7 A0A315V3X5 A0A0P5N7B8 G5BI84 H0VRB9 V9LD01 A0A1J4WNZ2 A0A2Y9GDT1 K9IQL2 A0A0L7R0U7 A0A091DB69 K1PBH8 A0A2L2Y921 A0A3B5MEY7 W5LZU0 A0A3B3SEY4 A0A0G0U9Y9 A0A0F8C880 L8Y3L9 A0A3B4X7C0 A0A2J7QAH4 P53368 A0A3L7HQ12 A0A3B4V3R6 A0A2Y9J9K4 A0A250YAK2 A0A226EFD6 A0A340Y2C8 A0A3B3B4G3 U6D127 A0A3Q4BXF9 A0A3Q7NCN7 A0A2U3VHB8 M3Z2J0 P53369 A0A2M7RHS1 T1J6Y5 A0A3Q1B3G7 A0A3P8THN9 A0A1S3JEB6 A0A2P4X9Z8 A0A2U9CT94 G1LAD2 F7IL36 A0A287AR30 A0A2U3XF28 H3CJ26 A0A1F7U7E9 A0A1D2MTM7 A0A3P8X2A5 W5P5K1 A0A2K6USY0 A0A067R9D0 A0A146XTG5 A0A1A6GSK5 A0A3B3YP22 A0A087UUR3 A0A146NAI6 A0A341CX57 A0A3P9JNP2 A0A024UP77 A0A2U4CBW9 A0A2Y9MQ33 A0A1U7T6Y9 A0A3B3I653 A0A384CNV9 A0A3Q3FRG5 A0A087YAZ4 A0A3B3VKL9
EC Number
3.6.1.55
Pubmed
19121390
26354079
22118469
28756777
26227816
23622113
+ More
18563158 28648823 21292972 29023486 20075255 21709235 21881562 17495919 28812685 29703783 21993625 21993624 24402279 27112493 22992520 26561354 29240929 25835551 23385571 7592783 9013634 16141072 15489334 11572992 21183079 21070968 29281266 29704459 28087693 29451363 7586133 11817101 28186564 20010809 15496914 27774985 27289101 24487278 20809919 24845553 17554307 24813606
18563158 28648823 21292972 29023486 20075255 21709235 21881562 17495919 28812685 29703783 21993625 21993624 24402279 27112493 22992520 26561354 29240929 25835551 23385571 7592783 9013634 16141072 15489334 11572992 21183079 21070968 29281266 29704459 28087693 29451363 7586133 11817101 28186564 20010809 15496914 27774985 27289101 24487278 20809919 24845553 17554307 24813606
EMBL
BABH01039827
KQ460044
KPJ18318.1
ODYU01012895
SOQ59429.1
AGBW02008849
+ More
OWR52370.1 KZ150306 PZC71498.1 NWSH01000621 PCG75247.1 JTDY01000984 KOB75198.1 KQ459232 KPJ02552.1 GAIX01006072 JAA86488.1 GG666633 EEN47234.1 AAGJ04013873 GDRN01010561 JAI67914.1 IACF01004779 LAB70368.1 LSMT01000020 PFX32675.1 HACG01039660 HACG01039661 CEK86525.1 CEK86526.1 NNAY01001222 OXU24692.1 GL732577 EFX75101.1 MRZV01001246 PIK39377.1 AEFK01076174 KE164528 EPQ18397.1 AGTP01110591 KQ435732 KOX77507.1 AAWZ02025450 PZQS01000001 PVD38768.1 GDIP01033361 LRGB01002060 JAM70354.1 KZS09539.1 KB108645 ELK28994.1 LK023346 CDS11240.1 NEDP02002059 OWF51812.1 NHOQ01002357 PWA17697.1 GDIQ01168236 JAK83489.1 JH170413 EHB08995.1 AAKN02032221 JW877372 AFP09889.1 MNWH01000042 OIO48961.1 GABZ01002805 JAA50720.1 KQ414669 KOC64474.1 KN124860 KFO20071.1 JH818586 EKC18833.1 IAAA01017335 LAA03830.1 AHAT01032352 LBYG01000005 KKR46962.1 KQ041428 KKF27055.1 KB366015 ELV11018.1 NEVH01016330 PNF25585.1 D49956 D88356 AK011695 AK088309 AK168312 CH466529 BC021940 BC098239 RAZU01000163 RLQ68027.1 GFFW01004194 JAV40594.1 LNIX01000004 OXA56262.1 HAAF01004835 CCP76660.1 AEYP01089007 D49977 D49980 PFME01000023 PIY96051.1 JH431897 NCKW01015590 POM62365.1 CP026261 AWP19180.1 ACTA01156832 AEMK02000013 MGEA01000052 OGL73657.1 LJIJ01000537 ODM96469.1 AMGL01069906 KK852804 KDR16209.1 GCES01040960 JAR45363.1 LZPO01073203 OBS69303.1 KK121735 KFM81102.1 GCES01158276 GCES01040959 JAQ28046.1 KI913953 ETW08261.1 AYCK01007272
OWR52370.1 KZ150306 PZC71498.1 NWSH01000621 PCG75247.1 JTDY01000984 KOB75198.1 KQ459232 KPJ02552.1 GAIX01006072 JAA86488.1 GG666633 EEN47234.1 AAGJ04013873 GDRN01010561 JAI67914.1 IACF01004779 LAB70368.1 LSMT01000020 PFX32675.1 HACG01039660 HACG01039661 CEK86525.1 CEK86526.1 NNAY01001222 OXU24692.1 GL732577 EFX75101.1 MRZV01001246 PIK39377.1 AEFK01076174 KE164528 EPQ18397.1 AGTP01110591 KQ435732 KOX77507.1 AAWZ02025450 PZQS01000001 PVD38768.1 GDIP01033361 LRGB01002060 JAM70354.1 KZS09539.1 KB108645 ELK28994.1 LK023346 CDS11240.1 NEDP02002059 OWF51812.1 NHOQ01002357 PWA17697.1 GDIQ01168236 JAK83489.1 JH170413 EHB08995.1 AAKN02032221 JW877372 AFP09889.1 MNWH01000042 OIO48961.1 GABZ01002805 JAA50720.1 KQ414669 KOC64474.1 KN124860 KFO20071.1 JH818586 EKC18833.1 IAAA01017335 LAA03830.1 AHAT01032352 LBYG01000005 KKR46962.1 KQ041428 KKF27055.1 KB366015 ELV11018.1 NEVH01016330 PNF25585.1 D49956 D88356 AK011695 AK088309 AK168312 CH466529 BC021940 BC098239 RAZU01000163 RLQ68027.1 GFFW01004194 JAV40594.1 LNIX01000004 OXA56262.1 HAAF01004835 CCP76660.1 AEYP01089007 D49977 D49980 PFME01000023 PIY96051.1 JH431897 NCKW01015590 POM62365.1 CP026261 AWP19180.1 ACTA01156832 AEMK02000013 MGEA01000052 OGL73657.1 LJIJ01000537 ODM96469.1 AMGL01069906 KK852804 KDR16209.1 GCES01040960 JAR45363.1 LZPO01073203 OBS69303.1 KK121735 KFM81102.1 GCES01158276 GCES01040959 JAQ28046.1 KI913953 ETW08261.1 AYCK01007272
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000037510
UP000053268
+ More
UP000001554 UP000007110 UP000225706 UP000215335 UP000005203 UP000000305 UP000230750 UP000002358 UP000007648 UP000005215 UP000053105 UP000001646 UP000245119 UP000002280 UP000192224 UP000076858 UP000242188 UP000006813 UP000005447 UP000248481 UP000053825 UP000028990 UP000005408 UP000261380 UP000018468 UP000261540 UP000011518 UP000261360 UP000235965 UP000000589 UP000273346 UP000261420 UP000248482 UP000198287 UP000265300 UP000261560 UP000261620 UP000286641 UP000245340 UP000000715 UP000002494 UP000257160 UP000265080 UP000085678 UP000246464 UP000008912 UP000008225 UP000008227 UP000245341 UP000007303 UP000094527 UP000265120 UP000002356 UP000233220 UP000027135 UP000092124 UP000261480 UP000054359 UP000265000 UP000252040 UP000265200 UP000024375 UP000245320 UP000248483 UP000189704 UP000001038 UP000261680 UP000291021 UP000261660 UP000028760 UP000261500
UP000001554 UP000007110 UP000225706 UP000215335 UP000005203 UP000000305 UP000230750 UP000002358 UP000007648 UP000005215 UP000053105 UP000001646 UP000245119 UP000002280 UP000192224 UP000076858 UP000242188 UP000006813 UP000005447 UP000248481 UP000053825 UP000028990 UP000005408 UP000261380 UP000018468 UP000261540 UP000011518 UP000261360 UP000235965 UP000000589 UP000273346 UP000261420 UP000248482 UP000198287 UP000265300 UP000261560 UP000261620 UP000286641 UP000245340 UP000000715 UP000002494 UP000257160 UP000265080 UP000085678 UP000246464 UP000008912 UP000008225 UP000008227 UP000245341 UP000007303 UP000094527 UP000265120 UP000002356 UP000233220 UP000027135 UP000092124 UP000261480 UP000054359 UP000265000 UP000252040 UP000265200 UP000024375 UP000245320 UP000248483 UP000189704 UP000001038 UP000261680 UP000291021 UP000261660 UP000028760 UP000261500
Interpro
Gene 3D
ProteinModelPortal
H9J857
A0A0N0PDS7
A0A2H1X285
A0A212FF71
A0A2W1B7M3
A0A2A4JT84
+ More
A0A0L7LI40 A0A194QAQ3 S4PB70 C3ZJU9 W4Z1F4 A0A0P4WJF1 A0A2P2I8N0 A0A2B4SW84 A0A0B7B346 A0A232F278 A0A088A8N2 E9GZ80 A0A2G8JUD6 K7IZ70 G3WX39 S7QBG4 I3MPS7 A0A0M9A593 H9GEQ8 A0A2T7PZD0 F7FR03 A0A1W4Y9D1 A0A0P6AKK3 L5LRE6 A0A077WVB4 A0A210QSX7 A0A315V3X5 A0A0P5N7B8 G5BI84 H0VRB9 V9LD01 A0A1J4WNZ2 A0A2Y9GDT1 K9IQL2 A0A0L7R0U7 A0A091DB69 K1PBH8 A0A2L2Y921 A0A3B5MEY7 W5LZU0 A0A3B3SEY4 A0A0G0U9Y9 A0A0F8C880 L8Y3L9 A0A3B4X7C0 A0A2J7QAH4 P53368 A0A3L7HQ12 A0A3B4V3R6 A0A2Y9J9K4 A0A250YAK2 A0A226EFD6 A0A340Y2C8 A0A3B3B4G3 U6D127 A0A3Q4BXF9 A0A3Q7NCN7 A0A2U3VHB8 M3Z2J0 P53369 A0A2M7RHS1 T1J6Y5 A0A3Q1B3G7 A0A3P8THN9 A0A1S3JEB6 A0A2P4X9Z8 A0A2U9CT94 G1LAD2 F7IL36 A0A287AR30 A0A2U3XF28 H3CJ26 A0A1F7U7E9 A0A1D2MTM7 A0A3P8X2A5 W5P5K1 A0A2K6USY0 A0A067R9D0 A0A146XTG5 A0A1A6GSK5 A0A3B3YP22 A0A087UUR3 A0A146NAI6 A0A341CX57 A0A3P9JNP2 A0A024UP77 A0A2U4CBW9 A0A2Y9MQ33 A0A1U7T6Y9 A0A3B3I653 A0A384CNV9 A0A3Q3FRG5 A0A087YAZ4 A0A3B3VKL9
A0A0L7LI40 A0A194QAQ3 S4PB70 C3ZJU9 W4Z1F4 A0A0P4WJF1 A0A2P2I8N0 A0A2B4SW84 A0A0B7B346 A0A232F278 A0A088A8N2 E9GZ80 A0A2G8JUD6 K7IZ70 G3WX39 S7QBG4 I3MPS7 A0A0M9A593 H9GEQ8 A0A2T7PZD0 F7FR03 A0A1W4Y9D1 A0A0P6AKK3 L5LRE6 A0A077WVB4 A0A210QSX7 A0A315V3X5 A0A0P5N7B8 G5BI84 H0VRB9 V9LD01 A0A1J4WNZ2 A0A2Y9GDT1 K9IQL2 A0A0L7R0U7 A0A091DB69 K1PBH8 A0A2L2Y921 A0A3B5MEY7 W5LZU0 A0A3B3SEY4 A0A0G0U9Y9 A0A0F8C880 L8Y3L9 A0A3B4X7C0 A0A2J7QAH4 P53368 A0A3L7HQ12 A0A3B4V3R6 A0A2Y9J9K4 A0A250YAK2 A0A226EFD6 A0A340Y2C8 A0A3B3B4G3 U6D127 A0A3Q4BXF9 A0A3Q7NCN7 A0A2U3VHB8 M3Z2J0 P53369 A0A2M7RHS1 T1J6Y5 A0A3Q1B3G7 A0A3P8THN9 A0A1S3JEB6 A0A2P4X9Z8 A0A2U9CT94 G1LAD2 F7IL36 A0A287AR30 A0A2U3XF28 H3CJ26 A0A1F7U7E9 A0A1D2MTM7 A0A3P8X2A5 W5P5K1 A0A2K6USY0 A0A067R9D0 A0A146XTG5 A0A1A6GSK5 A0A3B3YP22 A0A087UUR3 A0A146NAI6 A0A341CX57 A0A3P9JNP2 A0A024UP77 A0A2U4CBW9 A0A2Y9MQ33 A0A1U7T6Y9 A0A3B3I653 A0A384CNV9 A0A3Q3FRG5 A0A087YAZ4 A0A3B3VKL9
PDB
6EHH
E-value=6.68522e-40,
Score=407
Ontologies
GO
GO:0008413
GO:0006281
GO:0030246
GO:0046061
GO:0005829
GO:0042262
GO:0003924
GO:0005759
GO:0035539
GO:0005634
GO:0047693
GO:0006203
GO:0036219
GO:0030515
GO:0006195
GO:0005615
GO:0005886
GO:0001669
GO:0007568
GO:0031965
GO:0008584
GO:0005739
GO:0046686
GO:0005737
GO:0046872
GO:0016787
GO:0004553
GO:0006508
GO:0004672
GO:0030677
GO:0016155
GO:0003824
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus membrane
Cytoplasmic vesicle
Secretory vesicle
Acrosome
Nucleus
Nucleus membrane
Cytoplasmic vesicle
Secretory vesicle
Acrosome
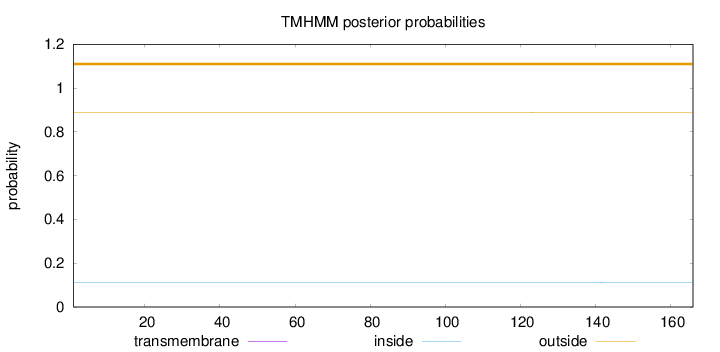
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01175
Exp number, first 60 AAs:
0.00031
Total prob of N-in:
0.11240
outside
1 - 166
Population Genetic Test Statistics
Pi
211.08525
Theta
183.920821
Tajima's D
0.536831
CLR
0.335894
CSRT
0.525923703814809
Interpretation
Uncertain
