Gene
KWMTBOMO10019
Pre Gene Modal
BGIBMGA005703
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.198
Sequence
CDS
ATGAAGTGTTTGAAGAACGGTTGCAGTGCTTACACCCAGGGTCCCGCAGGAGTGGCTTTTACAAATCTCATCACACACTTCCTAACCACACAATGTTTGATCACCGAGAACTGGCCTGAAGACAGATCTTACAGCTTAACAAATGGAGATATGTTCGATTTCATCATCGTTGGATCTGGTACTGCTGGTTCTCTACTGGCTAGCCGACTGACTGAAATTGAAGAGTGGAAGGTGCTGCTGATAGAAGCCGGTGGGGATCCTCCCTTAGAGAGTATTATTCCAAATTACTCTGGGGCAACACACCGAAGTTCTAGCGCGTTTCAGTATTACACTGAAATAGATGAGACCACTAATAAAGGAAGTCGCAACGAAAGGTCTTACTGGCCTCGCGGAAGAGTTCTCGGAGGAACAGGATCCATCAACGGCCTCCTCCATATGCGTGGCAGTCCGGAAGACTATAAGCCTTGGGCAGTAAACGAAGGTTGGGATTGGGAAACAATAAAAAAATATTTCATGAAGAGCGAGCGAATGATCGACCCTTTCATTCTCAACAATCCCGAACTAGCGAGAAGCCATGGCCTTGATGGTGAATTTGTGGTTGATCAATTAAACTTTACTCATTCTGAAATAGTTGATAAATTGACACAGGCTTATCAAGAGCTAGGCTTGAAATTTATAGATGATTTAAATGGAGATTCTCAAATGGGGGTGGGAAAGATTCGCGGCGGGAATCATAAAGGTAAAAGAGTTAGTACAGCATCAGCCTTCTTGAGTCCAATAAGAGAAAGAAAGAATTTGTACGTTTTGAAAAAAGCTTTTGCCAGGCATATAGATATAGAGCAGCCATCCAAAATAGCTAAAGGTGTAACAGTTTCTTTGAGTTATGGTAATGTAGAGACGTATTTTGCTAAAAAAGAAGTTATAGTAAGCGCAGGAGCAGTCAATACGCCAGCATTACTAATGATATCCGGTATAGGACCCCAAGAGCATTTTAAAGAACTGAAGTTTCTTAAACATTTAGTCAATCTACCTGTCGGAAGAAATTTACAAGATCATGTAAGGATACCTATACCAGTTACGCTTAATACTGGAGCCAAAGCAAAAACGGAGGAATATTGGCTTAAAGCTACAGCGGAGTACATTCTGCATCAAACAGGGCCTCTTGCTACTAATTACGATCAACCAAACATCAACGCCTTTCTTTCGGTACCTCATGGTAAAACTCAACCTGACGTGCAAATAGATCATAATTATTTCGTCCCAAACACATCCTATGTATTTACAATGTGTACAGAGATAATGTCATTTAAAGATGAAATTTGTCAGCAATTTAGTGATTTTAATACAGAAAAAGAAATGATAATATTTTTCGTGTCGCTTTGTAGGCCGCACTCGAGAGGTAAAATTGAGTTAAGAGGAATCAATGCAATGGAACATCCTAAAATATATTCAAAGTATTTTAGTGATGCACGTGATATGAAAACCTTTATTGGGGGGATAAAGAAAGTAACCGAGATAGTAAAAACACCAACGTTCCGTGATATGGACGCTAAATTAATGAGAATTAATTGGAGGGATTGTGATGGGTTTGAGTTTGAGAGTGACGAGTATTGGGAGTGCATGGCTAGAACTGTTACTTATAATGTTTATCATCCCGTCGGAACTGCTAAAATGGGGAAACCTAATGACCCAGAGGCAGTAGTTGACAATAAACTGAAGGTTTACAGTGTGAGAAATTTAAGAGTGGTAGACGCTAGCGTAATGCCTACAATACCCAGTGTCAATACAAATGCAGCTGTTATGATGATTGCTGAAAGAGCCGCTGATTTGATTAAAGAAGAATACGCAACTACAGTTAATACCATTAAAAAGGACGAATTGTAA
Protein
MKCLKNGCSAYTQGPAGVAFTNLITHFLTTQCLITENWPEDRSYSLTNGDMFDFIIVGSGTAGSLLASRLTEIEEWKVLLIEAGGDPPLESIIPNYSGATHRSSSAFQYYTEIDETTNKGSRNERSYWPRGRVLGGTGSINGLLHMRGSPEDYKPWAVNEGWDWETIKKYFMKSERMIDPFILNNPELARSHGLDGEFVVDQLNFTHSEIVDKLTQAYQELGLKFIDDLNGDSQMGVGKIRGGNHKGKRVSTASAFLSPIRERKNLYVLKKAFARHIDIEQPSKIAKGVTVSLSYGNVETYFAKKEVIVSAGAVNTPALLMISGIGPQEHFKELKFLKHLVNLPVGRNLQDHVRIPIPVTLNTGAKAKTEEYWLKATAEYILHQTGPLATNYDQPNINAFLSVPHGKTQPDVQIDHNYFVPNTSYVFTMCTEIMSFKDEICQQFSDFNTEKEMIIFFVSLCRPHSRGKIELRGINAMEHPKIYSKYFSDARDMKTFIGGIKKVTEIVKTPTFRDMDAKLMRINWRDCDGFEFESDEYWECMARTVTYNVYHPVGTAKMGKPNDPEAVVDNKLKVYSVRNLRVVDASVMPTIPSVNTNAAVMMIAERAADLIKEEYATTVNTIKKDEL
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9J861
A0A2A4IYS9
A0A2H1WZL6
A0A2W1B5P9
A0A0N1IJ07
A0A212FF54
+ More
A0A194QAU5 A0A0L7L582 H9JYB9 A0A194QB82 A0A2A4JLJ2 A0A1L8E0M0 A0A1L8E037 A0A2M4AFT0 A0A194QB47 A0A2M4AF63 A0A151K093 A0A2H1WG27 F4WRC9 A0A158NUQ9 A0A2J7PXM4 A0A195BBJ1 A0A182FZT6 A0A182IM41 A0A2M4ALF8 A0A1Y9H2C6 F5HJA8 F5HJA6 A0A1B0GIG3 A0A2J7RBI7 A0A182FZT4 A0A2M4AHF2 A0A151IVV0 A0A151J2Y2 H9JTY6 A0A2H1VF60 A0A2P8ZHS7 A0A182Q348 A0A2J7PXQ0 I6YR69 H9JTY5 A0A2J7QZ44 A0A2A4JKJ5 A0A182FZT5 A0A3S2P1X7 A0A2H1WT25 H9JTY7 A0A1B6LHN9 A0A2J7PXM0 Q7QFX9 A0A1Y9H2C4 A0A1L8E0E7 H9JTY8 A0A182GTK3 A0A0J7K353 A0A151WNR9 A0A182FZT3 A0A194PSS7 A0A067R8P7 A0A1B6EYD4 A0A1B6G282 A0A1L8DZX6 E0VUB2 A0A2P8YYA0 A0A084VW93 A0A1B6EWM9 A0A2P8ZGH8 A0A023EVK5 E2ACY0 A0A1B0D7C5 A0A0L7L9T7 F5HJA7 A0A1B0DMM3 A0A1B6HJ69 A0A2P8Z7X8 A0A1Y9H2V5 A0A336MWJ3 A0A2A4K4K5 A0A2A4JW72 A0A2H1VEJ4 A0A3L8D7F4 A0A067QT86 A0A154NWQ8 A0A2H1VTB0 T1PGK3 A0A1B6KJY4 A0A1I8MXV5 U5EVL2 A0A2H1V9N6 A0A2H1VDD9 B3NUX3 A0A088S376 A0A088SM07 T1H853 A0A2P8ZKS2 A0A2P8ZGP0 A0A1B6HLV3 A0A088S5A0 A0A088S3T2 A0A088S3M8
A0A194QAU5 A0A0L7L582 H9JYB9 A0A194QB82 A0A2A4JLJ2 A0A1L8E0M0 A0A1L8E037 A0A2M4AFT0 A0A194QB47 A0A2M4AF63 A0A151K093 A0A2H1WG27 F4WRC9 A0A158NUQ9 A0A2J7PXM4 A0A195BBJ1 A0A182FZT6 A0A182IM41 A0A2M4ALF8 A0A1Y9H2C6 F5HJA8 F5HJA6 A0A1B0GIG3 A0A2J7RBI7 A0A182FZT4 A0A2M4AHF2 A0A151IVV0 A0A151J2Y2 H9JTY6 A0A2H1VF60 A0A2P8ZHS7 A0A182Q348 A0A2J7PXQ0 I6YR69 H9JTY5 A0A2J7QZ44 A0A2A4JKJ5 A0A182FZT5 A0A3S2P1X7 A0A2H1WT25 H9JTY7 A0A1B6LHN9 A0A2J7PXM0 Q7QFX9 A0A1Y9H2C4 A0A1L8E0E7 H9JTY8 A0A182GTK3 A0A0J7K353 A0A151WNR9 A0A182FZT3 A0A194PSS7 A0A067R8P7 A0A1B6EYD4 A0A1B6G282 A0A1L8DZX6 E0VUB2 A0A2P8YYA0 A0A084VW93 A0A1B6EWM9 A0A2P8ZGH8 A0A023EVK5 E2ACY0 A0A1B0D7C5 A0A0L7L9T7 F5HJA7 A0A1B0DMM3 A0A1B6HJ69 A0A2P8Z7X8 A0A1Y9H2V5 A0A336MWJ3 A0A2A4K4K5 A0A2A4JW72 A0A2H1VEJ4 A0A3L8D7F4 A0A067QT86 A0A154NWQ8 A0A2H1VTB0 T1PGK3 A0A1B6KJY4 A0A1I8MXV5 U5EVL2 A0A2H1V9N6 A0A2H1VDD9 B3NUX3 A0A088S376 A0A088SM07 T1H853 A0A2P8ZKS2 A0A2P8ZGP0 A0A1B6HLV3 A0A088S5A0 A0A088S3T2 A0A088S3M8
Pubmed
EMBL
BABH01039828
NWSH01004668
PCG64789.1
ODYU01012241
SOQ58442.1
KZ150306
+ More
PZC71499.1 KQ460044 KPJ18321.1 AGBW02008849 OWR52369.1 KQ459232 KPJ02554.1 JTDY01002835 KOB70627.1 BABH01042896 KPJ02689.1 NWSH01001107 PCG72578.1 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 GGFK01006251 MBW39572.1 KPJ02687.1 GGFK01006105 MBW39426.1 KQ981296 KYN43077.1 ODYU01008368 SOQ51896.1 GL888284 EGI63346.1 ADTU01026683 NEVH01020853 PNF21087.1 KQ976532 KYM81564.1 GGFK01008304 MBW41625.1 AAAB01008844 EGK96369.1 EGK96367.1 EGK96368.1 AJWK01014080 NEVH01005904 PNF38181.1 GGFK01006898 MBW40219.1 KQ980881 KYN11981.1 KQ980314 KYN16689.1 BABH01003797 ODYU01002217 SOQ39416.1 PYGN01000053 PSN56054.1 AXCN02000040 PNF21103.1 JQ965926 AFN71166.1 NEVH01009076 PNF33851.1 PCG72577.1 RSAL01000051 RVE50264.1 ODYU01010827 SOQ56167.1 BABH01003798 BABH01003799 GEBQ01016745 JAT23232.1 PNF21089.1 EAA06000.3 GFDF01002042 JAV12042.1 JXUM01087318 KQ563625 KXJ73584.1 LBMM01015561 KMQ84762.1 KQ982905 KYQ49514.1 KQ459595 KPI96023.1 KK852811 KDR15959.1 GECZ01026787 JAS42982.1 GECZ01013224 JAS56545.1 GFDF01002041 JAV12043.1 DS235784 EEB16968.1 PYGN01000288 PSN49229.1 ATLV01017523 KE525172 KFB42237.1 GECZ01027417 JAS42352.1 PYGN01000064 PSN55598.1 GAPW01000526 JAC13072.1 GL438602 EFN68710.1 AJVK01026844 AJVK01026845 AJVK01026846 JTDY01002039 KOB72263.1 EGK96366.1 AJVK01016891 GECU01033045 JAS74661.1 PYGN01000155 PSN52601.1 UFQT01002442 SSX33469.1 NWSH01000190 PCG78592.1 NWSH01000521 PCG75884.1 ODYU01001942 SOQ38842.1 QOIP01000012 RLU16176.1 KK853558 KDR06741.1 KQ434773 KZC04023.1 ODYU01004023 SOQ43474.1 KA647270 AFP61899.1 GEBQ01028513 JAT11464.1 GANO01001829 JAB58042.1 ODYU01001405 SOQ37539.1 ODYU01001944 SOQ38845.1 CH954180 EDV47071.1 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 ACPB03005937 PYGN01000027 PSN57097.1 PSN55630.1 GECU01032039 JAS75667.1 KF717655 AIO02857.1 KF717661 AIO02863.1 KF717664 AIO02866.1
PZC71499.1 KQ460044 KPJ18321.1 AGBW02008849 OWR52369.1 KQ459232 KPJ02554.1 JTDY01002835 KOB70627.1 BABH01042896 KPJ02689.1 NWSH01001107 PCG72578.1 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 GGFK01006251 MBW39572.1 KPJ02687.1 GGFK01006105 MBW39426.1 KQ981296 KYN43077.1 ODYU01008368 SOQ51896.1 GL888284 EGI63346.1 ADTU01026683 NEVH01020853 PNF21087.1 KQ976532 KYM81564.1 GGFK01008304 MBW41625.1 AAAB01008844 EGK96369.1 EGK96367.1 EGK96368.1 AJWK01014080 NEVH01005904 PNF38181.1 GGFK01006898 MBW40219.1 KQ980881 KYN11981.1 KQ980314 KYN16689.1 BABH01003797 ODYU01002217 SOQ39416.1 PYGN01000053 PSN56054.1 AXCN02000040 PNF21103.1 JQ965926 AFN71166.1 NEVH01009076 PNF33851.1 PCG72577.1 RSAL01000051 RVE50264.1 ODYU01010827 SOQ56167.1 BABH01003798 BABH01003799 GEBQ01016745 JAT23232.1 PNF21089.1 EAA06000.3 GFDF01002042 JAV12042.1 JXUM01087318 KQ563625 KXJ73584.1 LBMM01015561 KMQ84762.1 KQ982905 KYQ49514.1 KQ459595 KPI96023.1 KK852811 KDR15959.1 GECZ01026787 JAS42982.1 GECZ01013224 JAS56545.1 GFDF01002041 JAV12043.1 DS235784 EEB16968.1 PYGN01000288 PSN49229.1 ATLV01017523 KE525172 KFB42237.1 GECZ01027417 JAS42352.1 PYGN01000064 PSN55598.1 GAPW01000526 JAC13072.1 GL438602 EFN68710.1 AJVK01026844 AJVK01026845 AJVK01026846 JTDY01002039 KOB72263.1 EGK96366.1 AJVK01016891 GECU01033045 JAS74661.1 PYGN01000155 PSN52601.1 UFQT01002442 SSX33469.1 NWSH01000190 PCG78592.1 NWSH01000521 PCG75884.1 ODYU01001942 SOQ38842.1 QOIP01000012 RLU16176.1 KK853558 KDR06741.1 KQ434773 KZC04023.1 ODYU01004023 SOQ43474.1 KA647270 AFP61899.1 GEBQ01028513 JAT11464.1 GANO01001829 JAB58042.1 ODYU01001405 SOQ37539.1 ODYU01001944 SOQ38845.1 CH954180 EDV47071.1 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 ACPB03005937 PYGN01000027 PSN57097.1 PSN55630.1 GECU01032039 JAS75667.1 KF717655 AIO02857.1 KF717661 AIO02863.1 KF717664 AIO02866.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000078541 UP000007755 UP000005205 UP000235965 UP000078540 UP000069272 UP000075880 UP000075884 UP000007062 UP000092461 UP000078492 UP000245037 UP000075886 UP000283053 UP000069940 UP000249989 UP000036403 UP000075809 UP000027135 UP000009046 UP000030765 UP000000311 UP000092462 UP000279307 UP000076502 UP000095301 UP000008711 UP000015103
UP000078541 UP000007755 UP000005205 UP000235965 UP000078540 UP000069272 UP000075880 UP000075884 UP000007062 UP000092461 UP000078492 UP000245037 UP000075886 UP000283053 UP000069940 UP000249989 UP000036403 UP000075809 UP000027135 UP000009046 UP000030765 UP000000311 UP000092462 UP000279307 UP000076502 UP000095301 UP000008711 UP000015103
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9J861
A0A2A4IYS9
A0A2H1WZL6
A0A2W1B5P9
A0A0N1IJ07
A0A212FF54
+ More
A0A194QAU5 A0A0L7L582 H9JYB9 A0A194QB82 A0A2A4JLJ2 A0A1L8E0M0 A0A1L8E037 A0A2M4AFT0 A0A194QB47 A0A2M4AF63 A0A151K093 A0A2H1WG27 F4WRC9 A0A158NUQ9 A0A2J7PXM4 A0A195BBJ1 A0A182FZT6 A0A182IM41 A0A2M4ALF8 A0A1Y9H2C6 F5HJA8 F5HJA6 A0A1B0GIG3 A0A2J7RBI7 A0A182FZT4 A0A2M4AHF2 A0A151IVV0 A0A151J2Y2 H9JTY6 A0A2H1VF60 A0A2P8ZHS7 A0A182Q348 A0A2J7PXQ0 I6YR69 H9JTY5 A0A2J7QZ44 A0A2A4JKJ5 A0A182FZT5 A0A3S2P1X7 A0A2H1WT25 H9JTY7 A0A1B6LHN9 A0A2J7PXM0 Q7QFX9 A0A1Y9H2C4 A0A1L8E0E7 H9JTY8 A0A182GTK3 A0A0J7K353 A0A151WNR9 A0A182FZT3 A0A194PSS7 A0A067R8P7 A0A1B6EYD4 A0A1B6G282 A0A1L8DZX6 E0VUB2 A0A2P8YYA0 A0A084VW93 A0A1B6EWM9 A0A2P8ZGH8 A0A023EVK5 E2ACY0 A0A1B0D7C5 A0A0L7L9T7 F5HJA7 A0A1B0DMM3 A0A1B6HJ69 A0A2P8Z7X8 A0A1Y9H2V5 A0A336MWJ3 A0A2A4K4K5 A0A2A4JW72 A0A2H1VEJ4 A0A3L8D7F4 A0A067QT86 A0A154NWQ8 A0A2H1VTB0 T1PGK3 A0A1B6KJY4 A0A1I8MXV5 U5EVL2 A0A2H1V9N6 A0A2H1VDD9 B3NUX3 A0A088S376 A0A088SM07 T1H853 A0A2P8ZKS2 A0A2P8ZGP0 A0A1B6HLV3 A0A088S5A0 A0A088S3T2 A0A088S3M8
A0A194QAU5 A0A0L7L582 H9JYB9 A0A194QB82 A0A2A4JLJ2 A0A1L8E0M0 A0A1L8E037 A0A2M4AFT0 A0A194QB47 A0A2M4AF63 A0A151K093 A0A2H1WG27 F4WRC9 A0A158NUQ9 A0A2J7PXM4 A0A195BBJ1 A0A182FZT6 A0A182IM41 A0A2M4ALF8 A0A1Y9H2C6 F5HJA8 F5HJA6 A0A1B0GIG3 A0A2J7RBI7 A0A182FZT4 A0A2M4AHF2 A0A151IVV0 A0A151J2Y2 H9JTY6 A0A2H1VF60 A0A2P8ZHS7 A0A182Q348 A0A2J7PXQ0 I6YR69 H9JTY5 A0A2J7QZ44 A0A2A4JKJ5 A0A182FZT5 A0A3S2P1X7 A0A2H1WT25 H9JTY7 A0A1B6LHN9 A0A2J7PXM0 Q7QFX9 A0A1Y9H2C4 A0A1L8E0E7 H9JTY8 A0A182GTK3 A0A0J7K353 A0A151WNR9 A0A182FZT3 A0A194PSS7 A0A067R8P7 A0A1B6EYD4 A0A1B6G282 A0A1L8DZX6 E0VUB2 A0A2P8YYA0 A0A084VW93 A0A1B6EWM9 A0A2P8ZGH8 A0A023EVK5 E2ACY0 A0A1B0D7C5 A0A0L7L9T7 F5HJA7 A0A1B0DMM3 A0A1B6HJ69 A0A2P8Z7X8 A0A1Y9H2V5 A0A336MWJ3 A0A2A4K4K5 A0A2A4JW72 A0A2H1VEJ4 A0A3L8D7F4 A0A067QT86 A0A154NWQ8 A0A2H1VTB0 T1PGK3 A0A1B6KJY4 A0A1I8MXV5 U5EVL2 A0A2H1V9N6 A0A2H1VDD9 B3NUX3 A0A088S376 A0A088SM07 T1H853 A0A2P8ZKS2 A0A2P8ZGP0 A0A1B6HLV3 A0A088S5A0 A0A088S3T2 A0A088S3M8
PDB
5NCC
E-value=4.734e-54,
Score=536
Ontologies
GO
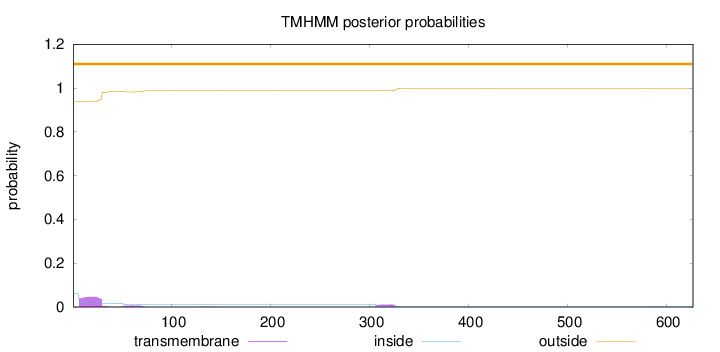
Topology
Length:
627
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.38359
Exp number, first 60 AAs:
1.08621
Total prob of N-in:
0.06093
outside
1 - 627
Population Genetic Test Statistics
Pi
293.21376
Theta
219.35122
Tajima's D
1.365303
CLR
0.143847
CSRT
0.754212289385531
Interpretation
Uncertain
