Gene
KWMTBOMO10006 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005695
Annotation
triacylglycerol_lipase_[Bombyx_mori]
Full name
Lipase
Location in the cell
Extracellular Reliability : 1.302 PlasmaMembrane Reliability : 1.341
Sequence
CDS
ATGAAGATGCACGTGTTCTTAGTTATTTTGGTGGCACTAGCGCGTGTCTCCGTCCAAGGGTTTGATTTGTTCGGAAGCTCGAGAAGCCTCGTCAATAACTTCGAAGACGCTCGTAACTACATCGAAACGCAAAAAGATAAGGTCATCGAAGAATGGTCCTCCTACATGGAAGACATAAAGCTGTCGAGCTCGTGGACGAACTTCTTAGAAGATAAAGAGAATCCGTCCTTGGAAGACCCCGACGTGGTGCTGAGCGTGCCCGCGATGATCACGCGGCGCGGCTACCGCTGTGAGACCCACTCGCTCATCTCCCAGGGATACGTGTTAAACATTCACAGGATACCGCAGGCTCGAAGCGGCGGCGACACTCCATCGAATACCGTGATACTTCAGCACGGCTTGTTTGCGAGCTCTGCCGACTGGGTCCTGAACGGGCCTGGAAAATCCTTAGCCTTCGTGTTGGCGGACGCCGGCTACGACGTCTGGATGCCAAACATAAGAGGTAACAGATATTCGAGGGAGCACACCACTTTAAAGAGCAGTTCGACTCAATATTGGAATTTCTCGTGGCACGAGGTCGCCCAGCACGACATCCCGGCCATCATAGACTATATCAGGGAGAGGAAGGGCTCGGACACAAAGATCGCGTACATGGGCCACTCGATGGGTTCCACGATGCTGTTCGCGATGCTGGCGCTGCGGCCGGAGTACAACGCGGTGCTGCGGGCGGGGCTCGCGCTCGGCCCCGTCGTCTACTTGTCTCATATCAAGTCGCCGGTCAAAACGCTAGCTCCGGTTGTCGCCAACGCGGCAAGAATGAACGTTATTAAAAACGGCGAATTTGTGCCGAAACAATCTGGCTTCGGACAAATGATGAGCGCGTGCAGCTCCGACGACGTGGACACGTACGTTTGTAAAAACGCAATATTCTTCATCTGCGGCACCGACGAGAAGCAGTTTAACAAGACGCTGCTGCCGGTGTTCCTGTCGCACCTCGGGACCGGCACGTCCATGAAGACCATCCTTCACTTCGCGCAGGAGATCGACGCCGCGGGACGCTTCCAGCAGTTCGACTACGGACCCACCAATAATATGAAAATATACAATTCAGAAACTCCACCAGAATACGATCTGCGCAAAATCACTCTTCCCATTTACCTGCTGTACTCAAGGAATGATTTATTGTCTAGCGAACAAGATGTTGATAAACTGTACCAAGATTTGGAAACCAGAACAGAAATCTACCTCGTACCGGACCCTGAGTTCAACCACGTGGATTACCTGATGGCGAACGACGCTCCGAGACTCTTGAACGACAAGGTGTTGCAGTTCTTGCTCGCAGCTTTTGCGGACCACCAAGGACCTGATCGTCACATGGGAACTCGCGAGTCAGTCCGCCAATACAAAGCATCACGTAAAGTGTTGAACACGTGA
Protein
MKMHVFLVILVALARVSVQGFDLFGSSRSLVNNFEDARNYIETQKDKVIEEWSSYMEDIKLSSSWTNFLEDKENPSLEDPDVVLSVPAMITRRGYRCETHSLISQGYVLNIHRIPQARSGGDTPSNTVILQHGLFASSADWVLNGPGKSLAFVLADAGYDVWMPNIRGNRYSREHTTLKSSSTQYWNFSWHEVAQHDIPAIIDYIRERKGSDTKIAYMGHSMGSTMLFAMLALRPEYNAVLRAGLALGPVVYLSHIKSPVKTLAPVVANAARMNVIKNGEFVPKQSGFGQMMSACSSDDVDTYVCKNAIFFICGTDEKQFNKTLLPVFLSHLGTGTSMKTILHFAQEIDAAGRFQQFDYGPTNNMKIYNSETPPEYDLRKITLPIYLLYSRNDLLSSEQDVDKLYQDLETRTEIYLVPDPEFNHVDYLMANDAPRLLNDKVLQFLLAAFADHQGPDRHMGTRESVRQYKASRKVLNT
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9J853
Q2F5V1
A0A0L7LD93
A0A2A4K5U1
S4NTM9
A0A2H1VNV1
+ More
A0A0N0PE67 A0A212EIJ7 A0A194QAJ7 A0A212FL02 A0A0N1IA21 A0A2A4K7B0 H9J5B8 A0A2W1BV18 A0A0N0P9P4 A0A2H1VB04 A0A0S1MM91 A0A0L7LBN2 A0A0L7KSX2 A0A1J1HYK0 A0A182YB52 A0A182NS05 A0A182FP77 A0A182M714 Q7PXY2 A0A182L9V7 A0A182JYW0 W5JQ53 A0A182HLG5 A0A084VGT2 A0A182PGK0 A0A182WSL5 A0A182VB02 A0A182TMH4 B0WDK6 A0A1Q3G4Y3 A0A2M4DRZ0 K7J2Y1 A0A182HBR9 A0A182RZA1 A0A182VZF7 A0A182INM5 Q16MC7 A0A1L8DQ12 A0A1L8DPQ6 A0A182G6M0 A0A067RCF7 A0A232EG55 A0A026X0Q1 A0A2M4AWW8 A0A2M4AXH6 E2B370 A0A2J7QW32 D2A3G9 A0A151X254 X1WSL9 A0A1B2M4Y7 A0A1B0CJ10 D2A0X3 A0A195CME3 V5FYL0 A0A0P4WII3 A0A1D2MRG2 A0A1W4W6G4 A0A195FK23 A0A1Y1KBU1 A0A226E1Q5 A0A0T6BEX7 A0A088A7F6 A0A1B6HSA1 A0A1Y1MRZ1 A0A1B6G3S2 A0A1S3CXK0 A0A194R2Y3 A0A2J7PY44 A0A151IQS0 A0A158NIA5 A0A194PL00 A0A310SHS6 A0A2P8XRN9 A0A336LYU0 A0A1B6C389 A0A154PPL4 A0A195AVW7 A0A1B0CTU8 A0A158P269 A0A067R0F9 V5GWI8 E2AHE1 N6TR89 U4U932 A0A2J7QG30 A0A195E3Z0 J3JU88 A0A195F6E3 A0A084W6W2 A0A139WJB3 A0A067QXT1 A0A139WJS1
A0A0N0PE67 A0A212EIJ7 A0A194QAJ7 A0A212FL02 A0A0N1IA21 A0A2A4K7B0 H9J5B8 A0A2W1BV18 A0A0N0P9P4 A0A2H1VB04 A0A0S1MM91 A0A0L7LBN2 A0A0L7KSX2 A0A1J1HYK0 A0A182YB52 A0A182NS05 A0A182FP77 A0A182M714 Q7PXY2 A0A182L9V7 A0A182JYW0 W5JQ53 A0A182HLG5 A0A084VGT2 A0A182PGK0 A0A182WSL5 A0A182VB02 A0A182TMH4 B0WDK6 A0A1Q3G4Y3 A0A2M4DRZ0 K7J2Y1 A0A182HBR9 A0A182RZA1 A0A182VZF7 A0A182INM5 Q16MC7 A0A1L8DQ12 A0A1L8DPQ6 A0A182G6M0 A0A067RCF7 A0A232EG55 A0A026X0Q1 A0A2M4AWW8 A0A2M4AXH6 E2B370 A0A2J7QW32 D2A3G9 A0A151X254 X1WSL9 A0A1B2M4Y7 A0A1B0CJ10 D2A0X3 A0A195CME3 V5FYL0 A0A0P4WII3 A0A1D2MRG2 A0A1W4W6G4 A0A195FK23 A0A1Y1KBU1 A0A226E1Q5 A0A0T6BEX7 A0A088A7F6 A0A1B6HSA1 A0A1Y1MRZ1 A0A1B6G3S2 A0A1S3CXK0 A0A194R2Y3 A0A2J7PY44 A0A151IQS0 A0A158NIA5 A0A194PL00 A0A310SHS6 A0A2P8XRN9 A0A336LYU0 A0A1B6C389 A0A154PPL4 A0A195AVW7 A0A1B0CTU8 A0A158P269 A0A067R0F9 V5GWI8 E2AHE1 N6TR89 U4U932 A0A2J7QG30 A0A195E3Z0 J3JU88 A0A195F6E3 A0A084W6W2 A0A139WJB3 A0A067QXT1 A0A139WJS1
Pubmed
EMBL
BABH01039868
KC702799
AB983536
AGN90922.1
BAQ20441.1
DQ311322
+ More
ABD36266.1 JTDY01001689 KOB73151.1 NWSH01000095 PCG79625.1 GAIX01012151 JAA80409.1 ODYU01003406 SOQ42132.1 KQ460044 KPJ18326.1 AGBW02014621 OWR41323.1 KQ459232 KPJ02558.1 AGBW02008987 AGBW02007925 OWR51966.1 OWR54421.1 KQ460426 KPJ14773.1 NWSH01000071 PCG79959.1 BABH01037038 KC702800 AGN90923.1 KZ149927 PZC77505.1 KQ459256 KPJ02070.1 ODYU01001573 SOQ37981.1 KR821071 ALL42058.1 JTDY01001814 KOB72805.1 JTDY01006004 KOB66378.1 CVRI01000027 CRK92404.1 AXCM01002120 AAAB01008987 EAA00922.5 ADMH02000799 ETN65025.1 APCN01000897 ATLV01013084 KE524839 KFB37176.1 DS231898 EDS44615.1 GFDL01000188 JAV34857.1 GGFL01016104 MBW80282.1 JXUM01125660 JXUM01125661 JXUM01125662 KQ566971 KXJ69694.1 CH477866 EAT35492.1 GFDF01005649 JAV08435.1 GFDF01005648 JAV08436.1 JXUM01045142 KQ561427 KXJ78623.1 KK852794 KDR16428.1 NNAY01004862 OXU17327.1 KK107054 EZA61581.1 GGFK01011976 MBW45297.1 GGFK01011977 MBW45298.1 GL445305 EFN89852.1 NEVH01009768 PNF32784.1 KQ971338 EFA02315.1 KQ982580 KYQ54458.1 ABLF02039332 KX447604 AOA60271.1 AJWK01013948 EFA01616.1 KQ977622 KYN01254.1 GALX01005701 JAB62765.1 GDRN01044880 JAI66895.1 LJIJ01000666 ODM95481.1 KQ981523 KYN40354.1 GEZM01086956 JAV58943.1 LNIX01000009 OXA50396.1 LJIG01001091 KRT85849.1 GECU01030185 JAS77521.1 GEZM01023396 JAV88411.1 GECZ01012738 JAS57031.1 KQ460855 KPJ11879.1 NEVH01020850 PNF21255.1 KQ976755 KYN08590.1 ADTU01016204 ADTU01016205 KQ459601 KPI94007.1 KQ759878 OAD62255.1 PYGN01001466 PSN34671.1 UFQT01000235 SSX22181.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 KQ435012 KZC13809.1 KQ976731 KYM76202.1 AJWK01028000 ADTU01007002 KK852827 KDR15388.1 GALX01003858 JAB64608.1 GL439509 EFN67149.1 APGK01057867 APGK01057868 KB741282 ENN70836.1 KB632233 ERL90414.1 NEVH01014836 PNF27554.1 KQ979657 KYN19900.1 BT126801 AEE61763.1 KQ981763 KYN36038.1 ATLV01020990 KE525311 KFB45956.1 KYB28090.1 KK853131 KDR10945.1 KYB28091.1
ABD36266.1 JTDY01001689 KOB73151.1 NWSH01000095 PCG79625.1 GAIX01012151 JAA80409.1 ODYU01003406 SOQ42132.1 KQ460044 KPJ18326.1 AGBW02014621 OWR41323.1 KQ459232 KPJ02558.1 AGBW02008987 AGBW02007925 OWR51966.1 OWR54421.1 KQ460426 KPJ14773.1 NWSH01000071 PCG79959.1 BABH01037038 KC702800 AGN90923.1 KZ149927 PZC77505.1 KQ459256 KPJ02070.1 ODYU01001573 SOQ37981.1 KR821071 ALL42058.1 JTDY01001814 KOB72805.1 JTDY01006004 KOB66378.1 CVRI01000027 CRK92404.1 AXCM01002120 AAAB01008987 EAA00922.5 ADMH02000799 ETN65025.1 APCN01000897 ATLV01013084 KE524839 KFB37176.1 DS231898 EDS44615.1 GFDL01000188 JAV34857.1 GGFL01016104 MBW80282.1 JXUM01125660 JXUM01125661 JXUM01125662 KQ566971 KXJ69694.1 CH477866 EAT35492.1 GFDF01005649 JAV08435.1 GFDF01005648 JAV08436.1 JXUM01045142 KQ561427 KXJ78623.1 KK852794 KDR16428.1 NNAY01004862 OXU17327.1 KK107054 EZA61581.1 GGFK01011976 MBW45297.1 GGFK01011977 MBW45298.1 GL445305 EFN89852.1 NEVH01009768 PNF32784.1 KQ971338 EFA02315.1 KQ982580 KYQ54458.1 ABLF02039332 KX447604 AOA60271.1 AJWK01013948 EFA01616.1 KQ977622 KYN01254.1 GALX01005701 JAB62765.1 GDRN01044880 JAI66895.1 LJIJ01000666 ODM95481.1 KQ981523 KYN40354.1 GEZM01086956 JAV58943.1 LNIX01000009 OXA50396.1 LJIG01001091 KRT85849.1 GECU01030185 JAS77521.1 GEZM01023396 JAV88411.1 GECZ01012738 JAS57031.1 KQ460855 KPJ11879.1 NEVH01020850 PNF21255.1 KQ976755 KYN08590.1 ADTU01016204 ADTU01016205 KQ459601 KPI94007.1 KQ759878 OAD62255.1 PYGN01001466 PSN34671.1 UFQT01000235 SSX22181.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 KQ435012 KZC13809.1 KQ976731 KYM76202.1 AJWK01028000 ADTU01007002 KK852827 KDR15388.1 GALX01003858 JAB64608.1 GL439509 EFN67149.1 APGK01057867 APGK01057868 KB741282 ENN70836.1 KB632233 ERL90414.1 NEVH01014836 PNF27554.1 KQ979657 KYN19900.1 BT126801 AEE61763.1 KQ981763 KYN36038.1 ATLV01020990 KE525311 KFB45956.1 KYB28090.1 KK853131 KDR10945.1 KYB28091.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000183832 UP000076408 UP000075884 UP000069272 UP000075883 UP000007062 UP000075882 UP000075881 UP000000673 UP000075840 UP000030765 UP000075885 UP000076407 UP000075903 UP000075902 UP000002320 UP000002358 UP000069940 UP000249989 UP000075900 UP000075920 UP000075880 UP000008820 UP000027135 UP000215335 UP000053097 UP000008237 UP000235965 UP000007266 UP000075809 UP000007819 UP000092461 UP000078542 UP000094527 UP000192223 UP000078541 UP000198287 UP000005203 UP000079169 UP000005205 UP000245037 UP000076502 UP000078540 UP000000311 UP000019118 UP000030742 UP000078492
UP000183832 UP000076408 UP000075884 UP000069272 UP000075883 UP000007062 UP000075882 UP000075881 UP000000673 UP000075840 UP000030765 UP000075885 UP000076407 UP000075903 UP000075902 UP000002320 UP000002358 UP000069940 UP000249989 UP000075900 UP000075920 UP000075880 UP000008820 UP000027135 UP000215335 UP000053097 UP000008237 UP000235965 UP000007266 UP000075809 UP000007819 UP000092461 UP000078542 UP000094527 UP000192223 UP000078541 UP000198287 UP000005203 UP000079169 UP000005205 UP000245037 UP000076502 UP000078540 UP000000311 UP000019118 UP000030742 UP000078492
Interpro
Gene 3D
ProteinModelPortal
H9J853
Q2F5V1
A0A0L7LD93
A0A2A4K5U1
S4NTM9
A0A2H1VNV1
+ More
A0A0N0PE67 A0A212EIJ7 A0A194QAJ7 A0A212FL02 A0A0N1IA21 A0A2A4K7B0 H9J5B8 A0A2W1BV18 A0A0N0P9P4 A0A2H1VB04 A0A0S1MM91 A0A0L7LBN2 A0A0L7KSX2 A0A1J1HYK0 A0A182YB52 A0A182NS05 A0A182FP77 A0A182M714 Q7PXY2 A0A182L9V7 A0A182JYW0 W5JQ53 A0A182HLG5 A0A084VGT2 A0A182PGK0 A0A182WSL5 A0A182VB02 A0A182TMH4 B0WDK6 A0A1Q3G4Y3 A0A2M4DRZ0 K7J2Y1 A0A182HBR9 A0A182RZA1 A0A182VZF7 A0A182INM5 Q16MC7 A0A1L8DQ12 A0A1L8DPQ6 A0A182G6M0 A0A067RCF7 A0A232EG55 A0A026X0Q1 A0A2M4AWW8 A0A2M4AXH6 E2B370 A0A2J7QW32 D2A3G9 A0A151X254 X1WSL9 A0A1B2M4Y7 A0A1B0CJ10 D2A0X3 A0A195CME3 V5FYL0 A0A0P4WII3 A0A1D2MRG2 A0A1W4W6G4 A0A195FK23 A0A1Y1KBU1 A0A226E1Q5 A0A0T6BEX7 A0A088A7F6 A0A1B6HSA1 A0A1Y1MRZ1 A0A1B6G3S2 A0A1S3CXK0 A0A194R2Y3 A0A2J7PY44 A0A151IQS0 A0A158NIA5 A0A194PL00 A0A310SHS6 A0A2P8XRN9 A0A336LYU0 A0A1B6C389 A0A154PPL4 A0A195AVW7 A0A1B0CTU8 A0A158P269 A0A067R0F9 V5GWI8 E2AHE1 N6TR89 U4U932 A0A2J7QG30 A0A195E3Z0 J3JU88 A0A195F6E3 A0A084W6W2 A0A139WJB3 A0A067QXT1 A0A139WJS1
A0A0N0PE67 A0A212EIJ7 A0A194QAJ7 A0A212FL02 A0A0N1IA21 A0A2A4K7B0 H9J5B8 A0A2W1BV18 A0A0N0P9P4 A0A2H1VB04 A0A0S1MM91 A0A0L7LBN2 A0A0L7KSX2 A0A1J1HYK0 A0A182YB52 A0A182NS05 A0A182FP77 A0A182M714 Q7PXY2 A0A182L9V7 A0A182JYW0 W5JQ53 A0A182HLG5 A0A084VGT2 A0A182PGK0 A0A182WSL5 A0A182VB02 A0A182TMH4 B0WDK6 A0A1Q3G4Y3 A0A2M4DRZ0 K7J2Y1 A0A182HBR9 A0A182RZA1 A0A182VZF7 A0A182INM5 Q16MC7 A0A1L8DQ12 A0A1L8DPQ6 A0A182G6M0 A0A067RCF7 A0A232EG55 A0A026X0Q1 A0A2M4AWW8 A0A2M4AXH6 E2B370 A0A2J7QW32 D2A3G9 A0A151X254 X1WSL9 A0A1B2M4Y7 A0A1B0CJ10 D2A0X3 A0A195CME3 V5FYL0 A0A0P4WII3 A0A1D2MRG2 A0A1W4W6G4 A0A195FK23 A0A1Y1KBU1 A0A226E1Q5 A0A0T6BEX7 A0A088A7F6 A0A1B6HSA1 A0A1Y1MRZ1 A0A1B6G3S2 A0A1S3CXK0 A0A194R2Y3 A0A2J7PY44 A0A151IQS0 A0A158NIA5 A0A194PL00 A0A310SHS6 A0A2P8XRN9 A0A336LYU0 A0A1B6C389 A0A154PPL4 A0A195AVW7 A0A1B0CTU8 A0A158P269 A0A067R0F9 V5GWI8 E2AHE1 N6TR89 U4U932 A0A2J7QG30 A0A195E3Z0 J3JU88 A0A195F6E3 A0A084W6W2 A0A139WJB3 A0A067QXT1 A0A139WJS1
PDB
1K8Q
E-value=3.13377e-65,
Score=631
Ontologies
Topology
Subcellular location
Cytoplasm
SignalP
Position: 1 - 20,
Likelihood: 0.996811
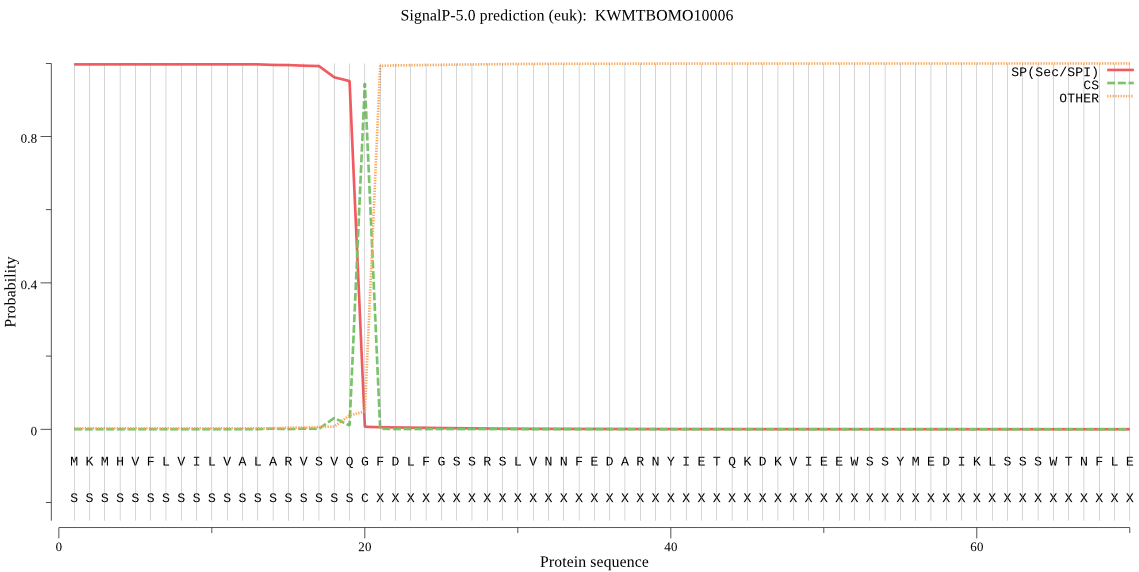
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.88571
Exp number, first 60 AAs:
1.65304
Total prob of N-in:
0.08791
outside
1 - 477

Population Genetic Test Statistics
Pi
227.752361
Theta
214.461897
Tajima's D
-1.394996
CLR
0.270457
CSRT
0.0764461776911154
Interpretation
Uncertain
