Gene
KWMTBOMO10005
Annotation
PREDICTED:_retrovirus-related_Pol_polyprotein_from_transposon_TNT_1-94_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.238 Extracellular Reliability : 1.125 Mitochondrial Reliability : 1.341
Sequence
CDS
ATGGGCCCAAATATGTTTAATTTAGAAAAACTGGTAGGAAGAGAAAACTTTGCCACGTGGAAATTCAGCATGAAAGCATATTTGGAACATGAAGACTTGTGGTGTTGTGTCGAATGCCACGACGGCAAGCCGGTGGATGCTGTAAAGGATATCAAAGCGAAATCAAAGCTGATCTTACTACTGGATCCGCAAAATTATGTCCATGTTCAAGACTGTAAAACCGTCAAACAAGTGTGGGAAAATTTGCAAAAGGCATTTGATGATAATGGTTTAACAAGAAGGGTAGGTTTGCTTAAAGATTTGATCAACACCACATTAGATTCAAGTAACAGAGTTGAAGATTACGTAAGTAAAATAATGAACACAGCACACAAGTTACGTAATATTAGTTTTGATGTCAATGATGAGTGGCTTGGCACCTTGATGTTGGCTGGCTTACCAGAAGAATACAAACCCATGATTATGGGCTTAGAGAGCTCTGGCATTAAAATAAGTGCTGATTCAGTAAAATCTAAACTTATACAAGAAGTTAATACATCAAAATCAGCAGCCTTCTATACAAATTCCAGAAAACCCAGAAATAATACCTCATCGATAAAAACAAAGGGTCCGAGATGCTTTAACTGCAATAGGAATGGACATTTCGCTAAATATTGTAAACTACCAAAGAAACAAACGGCAAATAAAGGAGAAAGTACTAGTTTTGTGGCAGCTTTTTCAGCTACAGTCAGCAAAAATGTAAGTAATGAATGGTACATTGACTCTGGTGCATCCATGCATATGACAAGCAGAATTGATTGGATGTATAATGTAACTGAATCATCTGTAAAAAACATCACTGTGGCTAACAGAGAACCCTTAGCTGTGAATGGAGTTGGCTGTGTGAACATTCAGCTAAGTCAAGAAAATAAAATACAAGTTAAAAATGTTTTATTTGTACCTGGGCTGGCTACTAATCTTCTCTCAGTTGGAGCTATTGTGAAAAATGGATATAAGGTCACATTTAATGACAAAGGATGTGAAGTTGGAAATAGTAAAGGTGAAGTAATATGCTCTGCAAAGCTGAAGAATAGTTTGTATGTATTAGAGACCTGTAAGGAAGTGGCCCACTTGGTCTCAAGAAATAAGAATAGCAATAATACTTACCTGTGGCACCTGCGGATGGGTCATTTGAATATGTCTGATGTAAAGAAGCTACCTACATGCTCTGAGGGAGTGACTCTTATTCCAGATGTAAATGATGTCACTTGCACACATTGTATGGAAGGTAAGCAAGCTAGGTTACCTTTTAAAAATACTGGAACAAGAGCTACGAGACCTCTAGAATTAATCCATTCTGACTTGTGTGGTCCTATGGAGAACATTTCATTTGGAGGATTTTAA
Protein
MGPNMFNLEKLVGRENFATWKFSMKAYLEHEDLWCCVECHDGKPVDAVKDIKAKSKLILLLDPQNYVHVQDCKTVKQVWENLQKAFDDNGLTRRVGLLKDLINTTLDSSNRVEDYVSKIMNTAHKLRNISFDVNDEWLGTLMLAGLPEEYKPMIMGLESSGIKISADSVKSKLIQEVNTSKSAAFYTNSRKPRNNTSSIKTKGPRCFNCNRNGHFAKYCKLPKKQTANKGESTSFVAAFSATVSKNVSNEWYIDSGASMHMTSRIDWMYNVTESSVKNITVANREPLAVNGVGCVNIQLSQENKIQVKNVLFVPGLATNLLSVGAIVKNGYKVTFNDKGCEVGNSKGEVICSAKLKNSLYVLETCKEVAHLVSRNKNSNNTYLWHLRMGHLNMSDVKKLPTCSEGVTLIPDVNDVTCTHCMEGKQARLPFKNTGTRATRPLELIHSDLCGPMENISFGGF
Summary
Uniprot
A0A2A4JRI1
A0A2A4J959
A0A194RU38
A0A194RJV9
A0A1Y1LWI3
H9JND2
+ More
A0A182GZK6 A0A0J7KCB5 A0A0J7N3H6 A0A0J7NHJ8 A0A0J7KAH3 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A224XA26 A0A0A9ZDS9 W4VRQ4 W4VRP9 W4VRP2 A0A1B6E9D0 A0A0A9XCL0 A0A1W7R670 A0A1W7R663 A0A0A9Z683 A0A0A9XEY5 A0A212F7V4 A0A0J7N4R7 A0A1W7R657 A0A1W7R6H6 A0A1W7R6J6 A0A1W7R6K6 A0A0A9XVJ9 A0A0A9YKS3 A0A0A9WPH5 A0A0A9WRE3 A0A0N1ICY1 A0A3L8DS90 A0A0J7JZN1 A0A0A1WXQ3 A0A0N1IFR1 A0A0J7K0M9 A0A146LFV8 A0A034VP70 A0A085MVP5 A0A0V0T127 A0A1E1WHM3 A0A0V0T2I4 A0A0V1FKH2 A0A085N572 A0A0V1APS7 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V0WQU6 A0A0V0TUP0 A0A0V0YJB3 A0A0A9WQD4 A0A0J7K5G8 K7JBY9 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2W1BXU7 A0A0V0WR33 A0A2J7R668 K7J5Z3 A0A0V0Z6Q0 A0A2H1WW51 V5GAG2 A0A0A9YFM0 A0A1Y3E9R1 A0A1Y1N3X2 E2A5G7 A0A0V0SAT7 A0A0V1LPQ0 A0A0K8U6S2 A0A0V0V098 A0A0V1NJN8 A0A0V1GS48 A0A0J7KG03 A0A0A9WJ72 A0A0A9WG89 A0A0V1FXR9 A0A0A1XLZ7 A0A251TN47
A0A182GZK6 A0A0J7KCB5 A0A0J7N3H6 A0A0J7NHJ8 A0A0J7KAH3 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A224XA26 A0A0A9ZDS9 W4VRQ4 W4VRP9 W4VRP2 A0A1B6E9D0 A0A0A9XCL0 A0A1W7R670 A0A1W7R663 A0A0A9Z683 A0A0A9XEY5 A0A212F7V4 A0A0J7N4R7 A0A1W7R657 A0A1W7R6H6 A0A1W7R6J6 A0A1W7R6K6 A0A0A9XVJ9 A0A0A9YKS3 A0A0A9WPH5 A0A0A9WRE3 A0A0N1ICY1 A0A3L8DS90 A0A0J7JZN1 A0A0A1WXQ3 A0A0N1IFR1 A0A0J7K0M9 A0A146LFV8 A0A034VP70 A0A085MVP5 A0A0V0T127 A0A1E1WHM3 A0A0V0T2I4 A0A0V1FKH2 A0A085N572 A0A0V1APS7 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V0WQU6 A0A0V0TUP0 A0A0V0YJB3 A0A0A9WQD4 A0A0J7K5G8 K7JBY9 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2W1BXU7 A0A0V0WR33 A0A2J7R668 K7J5Z3 A0A0V0Z6Q0 A0A2H1WW51 V5GAG2 A0A0A9YFM0 A0A1Y3E9R1 A0A1Y1N3X2 E2A5G7 A0A0V0SAT7 A0A0V1LPQ0 A0A0K8U6S2 A0A0V0V098 A0A0V1NJN8 A0A0V1GS48 A0A0J7KG03 A0A0A9WJ72 A0A0A9WG89 A0A0V1FXR9 A0A0A1XLZ7 A0A251TN47
Pubmed
EMBL
NWSH01000816
PCG74073.1
NWSH01002292
PCG68657.1
PCG68658.1
KQ459875
+ More
KPJ19636.1 KQ460297 KPJ16236.1 GEZM01045162 JAV77889.1 BABH01019436 JXUM01099713 KQ564513 KXJ72145.1 LBMM01009660 KMQ87962.1 LBMM01010768 KMQ87255.1 LBMM01004924 KMQ91985.1 LBMM01010791 KMQ87231.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 GFTR01008662 JAW07764.1 GBHO01003629 JAG39975.1 GANO01000510 JAB59361.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 GEDC01002834 JAS34464.1 GBHO01026223 JAG17381.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 GBHO01003630 JAG39974.1 GBHO01026221 JAG17383.1 AGBW02009826 OWR49812.1 LBMM01010129 KMQ87655.1 GEHC01000991 JAV46654.1 GEHC01000990 JAV46655.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 GBHO01020706 JAG22898.1 GBHO01011901 JAG31703.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 KQ460882 KPJ11482.1 QOIP01000004 RLU23300.1 LBMM01019465 KMQ83527.1 GBXI01011094 JAD03198.1 KQ460208 KPJ16606.1 LBMM01017716 KMQ83988.1 GDHC01012180 JAQ06449.1 GAKP01015352 JAC43600.1 KL367628 KFD61291.1 JYDJ01001086 KRX32699.1 GDQN01004560 JAT86494.1 JYDJ01000877 KRX33198.1 JYDT01000069 KRY86576.1 KL367553 KFD64618.1 JYDH01000330 KRY26802.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDJ01000808 KRX33413.1 JYDK01000071 KRX78123.1 JYDJ01000136 KRX42725.1 JYDU01000010 KRY00136.1 GBHO01036544 JAG07060.1 LBMM01013295 KMQ85718.1 AAZX01000798 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 KZ149913 PZC78057.1 KRX78146.1 NEVH01006980 PNF36330.1 JYDQ01000379 KRY07987.1 ODYU01011515 SOQ57290.1 GALX01001351 JAB67115.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 LVZM01023578 OUC39908.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 GL436933 EFN71322.1 JYDL01000021 KRX23821.1 JYDW01000017 KRZ61510.1 GDHF01030254 JAI22060.1 JYDN01000113 KRX56900.1 JYDM01000185 KRZ84108.1 JYDP01000350 KRZ01021.1 LBMM01008124 KMQ89126.1 GBHO01036148 JAG07456.1 GBHO01036147 JAG07457.1 JYDT01000024 KRY90099.1 GBXI01002679 JAD11613.1 CM007899 OTG12557.1
KPJ19636.1 KQ460297 KPJ16236.1 GEZM01045162 JAV77889.1 BABH01019436 JXUM01099713 KQ564513 KXJ72145.1 LBMM01009660 KMQ87962.1 LBMM01010768 KMQ87255.1 LBMM01004924 KMQ91985.1 LBMM01010791 KMQ87231.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 GFTR01008662 JAW07764.1 GBHO01003629 JAG39975.1 GANO01000510 JAB59361.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 GEDC01002834 JAS34464.1 GBHO01026223 JAG17381.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 GBHO01003630 JAG39974.1 GBHO01026221 JAG17383.1 AGBW02009826 OWR49812.1 LBMM01010129 KMQ87655.1 GEHC01000991 JAV46654.1 GEHC01000990 JAV46655.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 GBHO01020706 JAG22898.1 GBHO01011901 JAG31703.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 KQ460882 KPJ11482.1 QOIP01000004 RLU23300.1 LBMM01019465 KMQ83527.1 GBXI01011094 JAD03198.1 KQ460208 KPJ16606.1 LBMM01017716 KMQ83988.1 GDHC01012180 JAQ06449.1 GAKP01015352 JAC43600.1 KL367628 KFD61291.1 JYDJ01001086 KRX32699.1 GDQN01004560 JAT86494.1 JYDJ01000877 KRX33198.1 JYDT01000069 KRY86576.1 KL367553 KFD64618.1 JYDH01000330 KRY26802.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDJ01000808 KRX33413.1 JYDK01000071 KRX78123.1 JYDJ01000136 KRX42725.1 JYDU01000010 KRY00136.1 GBHO01036544 JAG07060.1 LBMM01013295 KMQ85718.1 AAZX01000798 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 KZ149913 PZC78057.1 KRX78146.1 NEVH01006980 PNF36330.1 JYDQ01000379 KRY07987.1 ODYU01011515 SOQ57290.1 GALX01001351 JAB67115.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 LVZM01023578 OUC39908.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 GL436933 EFN71322.1 JYDL01000021 KRX23821.1 JYDW01000017 KRZ61510.1 GDHF01030254 JAI22060.1 JYDN01000113 KRX56900.1 JYDM01000185 KRZ84108.1 JYDP01000350 KRZ01021.1 LBMM01008124 KMQ89126.1 GBHO01036148 JAG07456.1 GBHO01036147 JAG07457.1 JYDT01000024 KRY90099.1 GBXI01002679 JAD11613.1 CM007899 OTG12557.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000069940
UP000249989
UP000036403
+ More
UP000007151 UP000279307 UP000055048 UP000054995 UP000054776 UP000054805 UP000054826 UP000054632 UP000054673 UP000054815 UP000002358 UP000235965 UP000054783 UP000243006 UP000000311 UP000054630 UP000054721 UP000054681 UP000054924 UP000055024 UP000215914
UP000007151 UP000279307 UP000055048 UP000054995 UP000054776 UP000054805 UP000054826 UP000054632 UP000054673 UP000054815 UP000002358 UP000235965 UP000054783 UP000243006 UP000000311 UP000054630 UP000054721 UP000054681 UP000054924 UP000055024 UP000215914
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JRI1
A0A2A4J959
A0A194RU38
A0A194RJV9
A0A1Y1LWI3
H9JND2
+ More
A0A182GZK6 A0A0J7KCB5 A0A0J7N3H6 A0A0J7NHJ8 A0A0J7KAH3 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A224XA26 A0A0A9ZDS9 W4VRQ4 W4VRP9 W4VRP2 A0A1B6E9D0 A0A0A9XCL0 A0A1W7R670 A0A1W7R663 A0A0A9Z683 A0A0A9XEY5 A0A212F7V4 A0A0J7N4R7 A0A1W7R657 A0A1W7R6H6 A0A1W7R6J6 A0A1W7R6K6 A0A0A9XVJ9 A0A0A9YKS3 A0A0A9WPH5 A0A0A9WRE3 A0A0N1ICY1 A0A3L8DS90 A0A0J7JZN1 A0A0A1WXQ3 A0A0N1IFR1 A0A0J7K0M9 A0A146LFV8 A0A034VP70 A0A085MVP5 A0A0V0T127 A0A1E1WHM3 A0A0V0T2I4 A0A0V1FKH2 A0A085N572 A0A0V1APS7 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V0WQU6 A0A0V0TUP0 A0A0V0YJB3 A0A0A9WQD4 A0A0J7K5G8 K7JBY9 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2W1BXU7 A0A0V0WR33 A0A2J7R668 K7J5Z3 A0A0V0Z6Q0 A0A2H1WW51 V5GAG2 A0A0A9YFM0 A0A1Y3E9R1 A0A1Y1N3X2 E2A5G7 A0A0V0SAT7 A0A0V1LPQ0 A0A0K8U6S2 A0A0V0V098 A0A0V1NJN8 A0A0V1GS48 A0A0J7KG03 A0A0A9WJ72 A0A0A9WG89 A0A0V1FXR9 A0A0A1XLZ7 A0A251TN47
A0A182GZK6 A0A0J7KCB5 A0A0J7N3H6 A0A0J7NHJ8 A0A0J7KAH3 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A224XA26 A0A0A9ZDS9 W4VRQ4 W4VRP9 W4VRP2 A0A1B6E9D0 A0A0A9XCL0 A0A1W7R670 A0A1W7R663 A0A0A9Z683 A0A0A9XEY5 A0A212F7V4 A0A0J7N4R7 A0A1W7R657 A0A1W7R6H6 A0A1W7R6J6 A0A1W7R6K6 A0A0A9XVJ9 A0A0A9YKS3 A0A0A9WPH5 A0A0A9WRE3 A0A0N1ICY1 A0A3L8DS90 A0A0J7JZN1 A0A0A1WXQ3 A0A0N1IFR1 A0A0J7K0M9 A0A146LFV8 A0A034VP70 A0A085MVP5 A0A0V0T127 A0A1E1WHM3 A0A0V0T2I4 A0A0V1FKH2 A0A085N572 A0A0V1APS7 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V0WQU6 A0A0V0TUP0 A0A0V0YJB3 A0A0A9WQD4 A0A0J7K5G8 K7JBY9 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2W1BXU7 A0A0V0WR33 A0A2J7R668 K7J5Z3 A0A0V0Z6Q0 A0A2H1WW51 V5GAG2 A0A0A9YFM0 A0A1Y3E9R1 A0A1Y1N3X2 E2A5G7 A0A0V0SAT7 A0A0V1LPQ0 A0A0K8U6S2 A0A0V0V098 A0A0V1NJN8 A0A0V1GS48 A0A0J7KG03 A0A0A9WJ72 A0A0A9WG89 A0A0V1FXR9 A0A0A1XLZ7 A0A251TN47
Ontologies
KEGG
GO
PANTHER
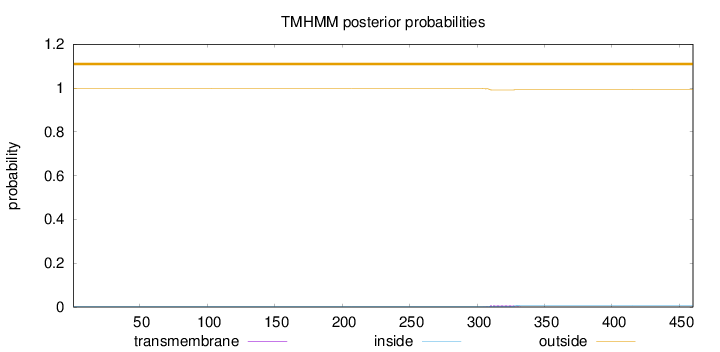
Topology
Length:
460
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15409
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00262
outside
1 - 460
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
