Gene
KWMTBOMO09992
Pre Gene Modal
BGIBMGA013207
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC103942885?_partial_[Pyrus_x_bretschneideri]
Location in the cell
Cytoplasmic Reliability : 1.319 Mitochondrial Reliability : 1.689
Sequence
CDS
ATGAGTAACAAGCCTCCGTTAAAACTGCAAAACGTTGATTTAGACTTCTTTACCGCAGAAACCCGGTCCAAATTAAATTCAATACTGGAAGCCTTCTGCTCCGTTTACCAGGCGGAGATGCAGGCTATTCTGAAAGCAACAGAAGTGATAGAGAGGCTTAAAGTAACAAAGGCAAACATCTTGAGCGACTCTAAATCATCTTTGGAAACATTGAAAGACCTCACCTCTCTCCACCCTGTCGCTTTCAGAATCAGAGCTAACATAGTGCGACTTGGGGCCGTCGGTGTGGACATCAAACTATTCTGGCCGAGGGCTCACGTAGGCAGCACTGCTGGCAACGAACGTGCGGATGAACCGGCGAAGACCGCTGCCCTGCACAGGAAAACTAAAGCTGTTTACAACTACTTTCCGCGCTCGTATGCTAGGAGGGTCATCAGGGAGGAAACGGTAACAAAATGGAATAAAGAATATAAACAGCTGACAAACAACAAGTCTATACAAACTTTCCTCCCTGATGTCCCTCTTGCATACATTTACGCAACAAACAATACACCTAACGGCATAGTAACACAAGTTTTAACGGGGCACGGAGGCTTTGGCCAATACTTACACCGCTTCAAGCTGCGTGAGAACCCGTGGTGCCCGTGTAAGCCTGGGGTAGTGGAAGACGTCCTGCATGTACTCCTGGAGTGCCCCAGATTCGGCGCTGCCCGGCAGCAGTTCGAGGCTGCAATCAACGACTCTCTGACGAGGGATCTGTTACACACTATTCTATAA
Protein
MSNKPPLKLQNVDLDFFTAETRSKLNSILEAFCSVYQAEMQAILKATEVIERLKVTKANILSDSKSSLETLKDLTSLHPVAFRIRANIVRLGAVGVDIKLFWPRAHVGSTAGNERADEPAKTAALHRKTKAVYNYFPRSYARRVIREETVTKWNKEYKQLTNNKSIQTFLPDVPLAYIYATNNTPNGIVTQVLTGHGGFGQYLHRFKLRENPWCPCKPGVVEDVLHVLLECPRFGAARQQFEAAINDSLTRDLLHTIL
Summary
Uniprot
H9JUJ4
A0A0L7KXZ2
A0A3S2TN34
A0A2W1BN41
A0A2A4JF48
A0A0L7L848
+ More
A0A0L7L1P2 Q9BLI5 Q3V5Y4 Q93139 A0A0L7LNK8 A0A437ART2 A0A0L7KPW9 A0A0K1IK14 A0A1W7R9Y4 A0A224XIA5 A0A437B2S6 A0A224X5M4 A0A0K1IK29 A0A437BVV1 A0A437BMS3 A0A224XI23 A0A2P8XKK4 A0A2L2YM05 A0A3S3P0A9 A0A087UJU4 A0A1W5CXT2 A0A0K8SHC4 A0A146MAU1 A0A0K8SHX4 A0A2P8YWT2 A0A3S2NBZ7 A0A087TCH8 O44317 A0A087TAD8 A0A224XJE3 A0A2E0U2U9 A0A087UUI6 A0A1V6PEQ8 A0A1V6PM44 B6H850 A0A1W4XBW8 A0A1W5CTD5 A0A0P4VS73 A0A1V6R2Q9 A0A2I4BSC2 A0A146QDM4 A0A1F5L1P6 A0A0A9VR33 A0A0U5GJ33 A0A146FYX2 A0A0K8SIU5 A0A438N3K9 A0A1F5L115 A0A0G4PXY0 A0A023EQ91 A0A131XRY9 A2QZW2 A0A146Q6P0 B6H8V5 A0A1W7R680 A0A146F6F4 A0A401L7X7 A0A401KR46 A0A1V6X7V7 A0A1F5L034 A0A147AN35 A0A254TW00 A0A1M3SYS3 A0A087T9L0 A0A293LVN9 G7Y200 F2S8R1 A0A1V6WQ47 A0A2P8Z0H0 A0A1V6X0H4 A0A1V6WRZ6 A0A1V6S8Z2 A0A1V6VN22 A0A087USZ4 A6R6U2 A0A1E3B1U8 A0A3Q0JKT9 A0A1B0GK10
A0A0L7L1P2 Q9BLI5 Q3V5Y4 Q93139 A0A0L7LNK8 A0A437ART2 A0A0L7KPW9 A0A0K1IK14 A0A1W7R9Y4 A0A224XIA5 A0A437B2S6 A0A224X5M4 A0A0K1IK29 A0A437BVV1 A0A437BMS3 A0A224XI23 A0A2P8XKK4 A0A2L2YM05 A0A3S3P0A9 A0A087UJU4 A0A1W5CXT2 A0A0K8SHC4 A0A146MAU1 A0A0K8SHX4 A0A2P8YWT2 A0A3S2NBZ7 A0A087TCH8 O44317 A0A087TAD8 A0A224XJE3 A0A2E0U2U9 A0A087UUI6 A0A1V6PEQ8 A0A1V6PM44 B6H850 A0A1W4XBW8 A0A1W5CTD5 A0A0P4VS73 A0A1V6R2Q9 A0A2I4BSC2 A0A146QDM4 A0A1F5L1P6 A0A0A9VR33 A0A0U5GJ33 A0A146FYX2 A0A0K8SIU5 A0A438N3K9 A0A1F5L115 A0A0G4PXY0 A0A023EQ91 A0A131XRY9 A2QZW2 A0A146Q6P0 B6H8V5 A0A1W7R680 A0A146F6F4 A0A401L7X7 A0A401KR46 A0A1V6X7V7 A0A1F5L034 A0A147AN35 A0A254TW00 A0A1M3SYS3 A0A087T9L0 A0A293LVN9 G7Y200 F2S8R1 A0A1V6WQ47 A0A2P8Z0H0 A0A1V6X0H4 A0A1V6WRZ6 A0A1V6S8Z2 A0A1V6VN22 A0A087USZ4 A6R6U2 A0A1E3B1U8 A0A3Q0JKT9 A0A1B0GK10
Pubmed
EMBL
BABH01022533
JTDY01004660
KOB67904.1
RSAL01000046
RVE50582.1
KZ149985
+ More
PZC75691.1 NWSH01001820 PCG70022.1 JTDY01002407 KOB71471.1 JTDY01003675 KOB69169.1 AB046668 BAB21511.1 AB211537 BAE44464.1 D38414 BAA07467.1 JTDY01000474 KOB77015.1 RSAL01001990 RVE40611.1 JTDY01007234 KOB65358.1 KP771712 AKU04650.1 GFAH01000444 JAV47945.1 GFTR01004219 JAW12207.1 RSAL01000192 RVE44624.1 GFTR01008639 JAW07787.1 KP771711 AKU04648.1 RSAL01000004 RVE54544.1 RSAL01000031 RVE51585.1 GFTR01004280 JAW12146.1 PYGN01001848 PSN32539.1 IAAA01038678 LAA08500.1 NCKU01007943 RWS02258.1 KK120149 KFM77633.1 FWEW01000736 SLM35555.1 GBRD01013108 JAG52718.1 GDHC01002702 JAQ15927.1 GBRD01013107 JAG52719.1 PYGN01000313 PSN48706.1 RSAL01000249 RVE43511.1 KK114580 KFM62817.1 AF015813 AAB94039.1 KK114278 KFM62077.1 GFTR01003820 JAW12606.1 PABS01000003 MAT33547.1 KK121687 KFM81025.1 MDYN01000160 OQD75006.1 MDYN01000090 OQD78085.1 AM920431 CAP93579.1 FWEW01000197 SLM33995.1 GDKW01003718 JAI52877.1 MDYP01000108 OQD95557.1 GCES01132138 JAQ54184.1 LXJU01000061 OGE47125.1 GBHO01044712 JAF98891.1 CDMC01000034 CEL11878.1 BCWF01000030 GAT29901.1 GBRD01013106 JAG52720.1 NAJM01000024 RVX70312.1 LXJU01000093 OGE46892.1 HG793200 CRL30999.1 GAPW01002368 JAC11230.1 GEFM01006389 JAP69407.1 AM270275 CAK41174.1 GCES01134373 JAQ51949.1 CAP93597.1 GEHC01000979 JAV46666.1 BCWF01000012 GAT21874.1 BDHI01000029 GCB27624.1 BDHI01000008 GCB21760.1 MOOB01000212 MOOB01000209 MOOB01000109 MOOB01000066 MOOB01000048 OQE65361.1 OQE65447.1 OQE71176.1 OQE76607.1 OQE80121.1 LXJU01000208 OGE46524.1 GCES01006353 JAR79970.1 OGUI01000013 SPB50616.1 KV878279 OJZ79659.1 KK114163 KFM61799.1 GFWV01007108 MAA31838.1 DF126536 GAA93209.1 GG698529 EGD99960.1 MOOB01000219 OQE64972.1 PYGN01000254 PSN49993.1 MOOB01000149 OQE68657.1 MOOB01000203 OQE65656.1 MLQL01000095 OQE10083.1 MOOB01000493 OQE52042.1 KK121445 KFM80483.1 CH476659 EDN08851.1 JXNT01000020 ODM14912.1 AJWK01022496 AJWK01022497
PZC75691.1 NWSH01001820 PCG70022.1 JTDY01002407 KOB71471.1 JTDY01003675 KOB69169.1 AB046668 BAB21511.1 AB211537 BAE44464.1 D38414 BAA07467.1 JTDY01000474 KOB77015.1 RSAL01001990 RVE40611.1 JTDY01007234 KOB65358.1 KP771712 AKU04650.1 GFAH01000444 JAV47945.1 GFTR01004219 JAW12207.1 RSAL01000192 RVE44624.1 GFTR01008639 JAW07787.1 KP771711 AKU04648.1 RSAL01000004 RVE54544.1 RSAL01000031 RVE51585.1 GFTR01004280 JAW12146.1 PYGN01001848 PSN32539.1 IAAA01038678 LAA08500.1 NCKU01007943 RWS02258.1 KK120149 KFM77633.1 FWEW01000736 SLM35555.1 GBRD01013108 JAG52718.1 GDHC01002702 JAQ15927.1 GBRD01013107 JAG52719.1 PYGN01000313 PSN48706.1 RSAL01000249 RVE43511.1 KK114580 KFM62817.1 AF015813 AAB94039.1 KK114278 KFM62077.1 GFTR01003820 JAW12606.1 PABS01000003 MAT33547.1 KK121687 KFM81025.1 MDYN01000160 OQD75006.1 MDYN01000090 OQD78085.1 AM920431 CAP93579.1 FWEW01000197 SLM33995.1 GDKW01003718 JAI52877.1 MDYP01000108 OQD95557.1 GCES01132138 JAQ54184.1 LXJU01000061 OGE47125.1 GBHO01044712 JAF98891.1 CDMC01000034 CEL11878.1 BCWF01000030 GAT29901.1 GBRD01013106 JAG52720.1 NAJM01000024 RVX70312.1 LXJU01000093 OGE46892.1 HG793200 CRL30999.1 GAPW01002368 JAC11230.1 GEFM01006389 JAP69407.1 AM270275 CAK41174.1 GCES01134373 JAQ51949.1 CAP93597.1 GEHC01000979 JAV46666.1 BCWF01000012 GAT21874.1 BDHI01000029 GCB27624.1 BDHI01000008 GCB21760.1 MOOB01000212 MOOB01000209 MOOB01000109 MOOB01000066 MOOB01000048 OQE65361.1 OQE65447.1 OQE71176.1 OQE76607.1 OQE80121.1 LXJU01000208 OGE46524.1 GCES01006353 JAR79970.1 OGUI01000013 SPB50616.1 KV878279 OJZ79659.1 KK114163 KFM61799.1 GFWV01007108 MAA31838.1 DF126536 GAA93209.1 GG698529 EGD99960.1 MOOB01000219 OQE64972.1 PYGN01000254 PSN49993.1 MOOB01000149 OQE68657.1 MOOB01000203 OQE65656.1 MLQL01000095 OQE10083.1 MOOB01000493 OQE52042.1 KK121445 KFM80483.1 CH476659 EDN08851.1 JXNT01000020 ODM14912.1 AJWK01022496 AJWK01022497
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000245037
UP000285301
+ More
UP000054359 UP000192927 UP000191672 UP000000724 UP000192223 UP000191518 UP000192220 UP000177622 UP000054771 UP000075230 UP000288859 UP000053732 UP000006706 UP000286921 UP000191691 UP000236662 UP000184063 UP000006812 UP000009172 UP000191342 UP000009297 UP000094569 UP000079169 UP000092461
UP000054359 UP000192927 UP000191672 UP000000724 UP000192223 UP000191518 UP000192220 UP000177622 UP000054771 UP000075230 UP000288859 UP000053732 UP000006706 UP000286921 UP000191691 UP000236662 UP000184063 UP000006812 UP000009172 UP000191342 UP000009297 UP000094569 UP000079169 UP000092461
Pfam
Interpro
IPR002156
RNaseH_domain
+ More
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR038902 INTS1
IPR040167 TF_CP2-like
IPR007604 CP2
IPR016035 Acyl_Trfase/lysoPLipase
IPR011990 TPR-like_helical_dom_sf
IPR002151 Kinesin_light
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR002182 NB-ARC
IPR027417 P-loop_NTPase
IPR031352 SesA
IPR001878 Znf_CCHC
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR038902 INTS1
IPR040167 TF_CP2-like
IPR007604 CP2
IPR016035 Acyl_Trfase/lysoPLipase
IPR011990 TPR-like_helical_dom_sf
IPR002151 Kinesin_light
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR002182 NB-ARC
IPR027417 P-loop_NTPase
IPR031352 SesA
IPR001878 Znf_CCHC
SUPFAM
Gene 3D
ProteinModelPortal
H9JUJ4
A0A0L7KXZ2
A0A3S2TN34
A0A2W1BN41
A0A2A4JF48
A0A0L7L848
+ More
A0A0L7L1P2 Q9BLI5 Q3V5Y4 Q93139 A0A0L7LNK8 A0A437ART2 A0A0L7KPW9 A0A0K1IK14 A0A1W7R9Y4 A0A224XIA5 A0A437B2S6 A0A224X5M4 A0A0K1IK29 A0A437BVV1 A0A437BMS3 A0A224XI23 A0A2P8XKK4 A0A2L2YM05 A0A3S3P0A9 A0A087UJU4 A0A1W5CXT2 A0A0K8SHC4 A0A146MAU1 A0A0K8SHX4 A0A2P8YWT2 A0A3S2NBZ7 A0A087TCH8 O44317 A0A087TAD8 A0A224XJE3 A0A2E0U2U9 A0A087UUI6 A0A1V6PEQ8 A0A1V6PM44 B6H850 A0A1W4XBW8 A0A1W5CTD5 A0A0P4VS73 A0A1V6R2Q9 A0A2I4BSC2 A0A146QDM4 A0A1F5L1P6 A0A0A9VR33 A0A0U5GJ33 A0A146FYX2 A0A0K8SIU5 A0A438N3K9 A0A1F5L115 A0A0G4PXY0 A0A023EQ91 A0A131XRY9 A2QZW2 A0A146Q6P0 B6H8V5 A0A1W7R680 A0A146F6F4 A0A401L7X7 A0A401KR46 A0A1V6X7V7 A0A1F5L034 A0A147AN35 A0A254TW00 A0A1M3SYS3 A0A087T9L0 A0A293LVN9 G7Y200 F2S8R1 A0A1V6WQ47 A0A2P8Z0H0 A0A1V6X0H4 A0A1V6WRZ6 A0A1V6S8Z2 A0A1V6VN22 A0A087USZ4 A6R6U2 A0A1E3B1U8 A0A3Q0JKT9 A0A1B0GK10
A0A0L7L1P2 Q9BLI5 Q3V5Y4 Q93139 A0A0L7LNK8 A0A437ART2 A0A0L7KPW9 A0A0K1IK14 A0A1W7R9Y4 A0A224XIA5 A0A437B2S6 A0A224X5M4 A0A0K1IK29 A0A437BVV1 A0A437BMS3 A0A224XI23 A0A2P8XKK4 A0A2L2YM05 A0A3S3P0A9 A0A087UJU4 A0A1W5CXT2 A0A0K8SHC4 A0A146MAU1 A0A0K8SHX4 A0A2P8YWT2 A0A3S2NBZ7 A0A087TCH8 O44317 A0A087TAD8 A0A224XJE3 A0A2E0U2U9 A0A087UUI6 A0A1V6PEQ8 A0A1V6PM44 B6H850 A0A1W4XBW8 A0A1W5CTD5 A0A0P4VS73 A0A1V6R2Q9 A0A2I4BSC2 A0A146QDM4 A0A1F5L1P6 A0A0A9VR33 A0A0U5GJ33 A0A146FYX2 A0A0K8SIU5 A0A438N3K9 A0A1F5L115 A0A0G4PXY0 A0A023EQ91 A0A131XRY9 A2QZW2 A0A146Q6P0 B6H8V5 A0A1W7R680 A0A146F6F4 A0A401L7X7 A0A401KR46 A0A1V6X7V7 A0A1F5L034 A0A147AN35 A0A254TW00 A0A1M3SYS3 A0A087T9L0 A0A293LVN9 G7Y200 F2S8R1 A0A1V6WQ47 A0A2P8Z0H0 A0A1V6X0H4 A0A1V6WRZ6 A0A1V6S8Z2 A0A1V6VN22 A0A087USZ4 A6R6U2 A0A1E3B1U8 A0A3Q0JKT9 A0A1B0GK10
Ontologies
GO
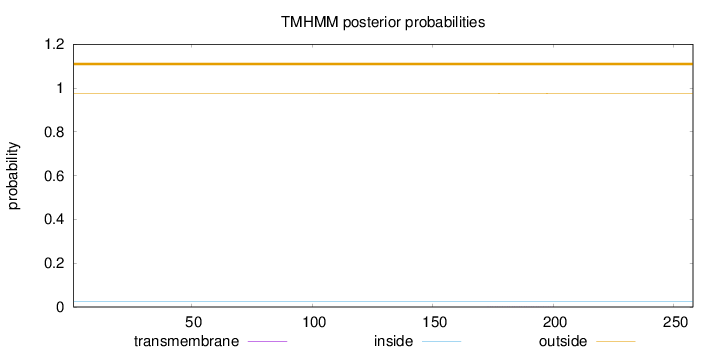
Topology
Length:
258
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01512
Exp number, first 60 AAs:
0.0006
Total prob of N-in:
0.02388
outside
1 - 258
Population Genetic Test Statistics
Pi
185.6111
Theta
174.184116
Tajima's D
0.251312
CLR
0.164603
CSRT
0.43902804859757
Interpretation
Uncertain
