Gene
KWMTBOMO09986
Annotation
Yokozuna_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.592 Nuclear Reliability : 2.152
Sequence
CDS
ATGTTGGGATATAGTAGGCGAAATGGAATAATACTTCAATTTACACAGCCATACTCACCGCAGTCCAACGGAATTGCAGAAAGAATGAATAGGACAATATACAATAAAGCAAGGACTATACTAAATGAGACAAGACTACCAAAACATCTATGGGGTGAAGCTGTATATCAAATAAACAGATGTCCATCATCAGCAATAAAGAATAAAGTACCTGCAGCAATAATGTTTAGAAAATTGGACTTATCAAAACTGCGAGTATTCGGTTCAAAAGTATGGGTGACTAATGTACCAAAGCAAGAGAAATTTGAAAATAGAGCAAAAGAAATGAGAATGGTTGGATATACACCAATTGGATGGAGACTATGGGATCCTAAAGAAAATATAATAGTATGCTTAAGAGATGTAAGATTTGATGAAAATGACTTAAAATATGAAGAGAATAAACTAAAGCAAACTGAAATAGAAGAAACTGTTATCTACAAGGATAAAGAAGAAGAAACTCACCTAGAAGATGAAGAGAAAATATTAAGAGAAGATGAAAATAAAGAAAGAAAGAGAGAACCTACTAAATATCAAGAAGCAGTACAAGTCAAAGAATGGCGACAAGCTATAAATAAAGAACTTGATTCACTTAATAAATTGAAAACATGGGAAGAAACAGAACTACCAAAAGGACAAAAAGCTATAGATACCAAGTGGATATTTAGAATGAAAGAAGATGGAACAAAGAAAGCAACAACAAGGACAAGAGGGATACTATGCACCAGTAGCAAGAATGTCAACAATACGAATTATGCTAATGCAGAAAAAGATGAAAATATTGATGTACCATACAAAGAACTTATTGGTTCATTACTTTTTATAGCAATGAACACTAGACCAGATATCTGTTATCCAGTATCATATCTGAGTAGATTTATGGACAACCCAACAACATCTTTGTGGAAAGCTGCAAAGATAGTACTTCGATATTTAAAATGCACACAAAATAAACCTTAA
Protein
MLGYSRRNGIILQFTQPYSPQSNGIAERMNRTIYNKARTILNETRLPKHLWGEAVYQINRCPSSAIKNKVPAAIMFRKLDLSKLRVFGSKVWVTNVPKQEKFENRAKEMRMVGYTPIGWRLWDPKENIIVCLRDVRFDENDLKYEENKLKQTEIEETVIYKDKEEETHLEDEEKILREDENKERKREPTKYQEAVQVKEWRQAINKELDSLNKLKTWEETELPKGQKAIDTKWIFRMKEDGTKKATTRTRGILCTSSKNVNNTNYANAEKDENIDVPYKELIGSLLFIAMNTRPDICYPVSYLSRFMDNPTTSLWKAAKIVLRYLKCTQNKP
Summary
Similarity
Belongs to the disease resistance NB-LRR family.
Belongs to the terpene synthase family.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Belongs to the terpene synthase family.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Uniprot
A0A0A9YSA9
A0A0A9YQT1
A0A0A9WQU6
A0A0A9Z0I6
A0A146KTS1
A0A0A9XG27
+ More
A0A2Z6MRL3 A0A0A9ZBW6 J9M5U1 J9LXN4 A0A0T6AT79 B8YLY3 X1WUQ9 B8YLY6 A0A1B6FK59 B8YLY5 A0A438DPU0 B8YLY4 A0A151T1X7 A0A1Y1L115 A0A139W9T1 A0A438F3A5 A0A438JWX0 A0A2S2PNK0 Q9C536 A5BMR0 A5BR97 A5BPR9 A0A151TJG4 A0A438CN15 A0A438FT52 B8YLY7 A0A438DEP9 A5B9M8 A0A0A9X1V9 A0A023F102 A0A0J7N374 A0A438CAN0 A0A2J7QL79 A0A2Z6PI76 A0A1U8HMK8 A0A182HBD9 A0A438E1L6 A0A1Y1MP14 A0A2U1MK50 A0A2Z6NYP5 A0A1Y1MQF0 A0A2N9HZS3 A0A1B6C2D3 A0A2N9I807 A0A2N9H4R5 A0A2N9FPD9 A0A2N9FL83 A0A2N9HZR4 A0A146KP10 A0A2N9FGL6 A0A2N9GG47 A0A1U8JW41 A0A2N9HHJ9 A0A2N9J203 A0A2N9HY48 A0A2U1M839 A0A2K3N600 A0A2N9EER7 A0A2N9G604 A0A2N9I9F7 H3H993 A0A396INR0 A0A438HY27 A5AFU8 A5B0R8 A0A0J7K689 A0A2Z6N7E5 A0A2N9I6M8 A0A2N9I0K6 A5BBA3 A0A445G8E4 A0A2N9IGN4 A0A2N9H7C1 A0A2N9IR42 A0A2N9IGI9 A0A1Y3E9R1
A0A2Z6MRL3 A0A0A9ZBW6 J9M5U1 J9LXN4 A0A0T6AT79 B8YLY3 X1WUQ9 B8YLY6 A0A1B6FK59 B8YLY5 A0A438DPU0 B8YLY4 A0A151T1X7 A0A1Y1L115 A0A139W9T1 A0A438F3A5 A0A438JWX0 A0A2S2PNK0 Q9C536 A5BMR0 A5BR97 A5BPR9 A0A151TJG4 A0A438CN15 A0A438FT52 B8YLY7 A0A438DEP9 A5B9M8 A0A0A9X1V9 A0A023F102 A0A0J7N374 A0A438CAN0 A0A2J7QL79 A0A2Z6PI76 A0A1U8HMK8 A0A182HBD9 A0A438E1L6 A0A1Y1MP14 A0A2U1MK50 A0A2Z6NYP5 A0A1Y1MQF0 A0A2N9HZS3 A0A1B6C2D3 A0A2N9I807 A0A2N9H4R5 A0A2N9FPD9 A0A2N9FL83 A0A2N9HZR4 A0A146KP10 A0A2N9FGL6 A0A2N9GG47 A0A1U8JW41 A0A2N9HHJ9 A0A2N9J203 A0A2N9HY48 A0A2U1M839 A0A2K3N600 A0A2N9EER7 A0A2N9G604 A0A2N9I9F7 H3H993 A0A396INR0 A0A438HY27 A5AFU8 A5B0R8 A0A0J7K689 A0A2Z6N7E5 A0A2N9I6M8 A0A2N9I0K6 A5BBA3 A0A445G8E4 A0A2N9IGN4 A0A2N9H7C1 A0A2N9IR42 A0A2N9IGI9 A0A1Y3E9R1
Pubmed
EMBL
GBHO01009621
JAG33983.1
GBHO01009623
JAG33981.1
GBHO01034731
JAG08873.1
+ More
GBHO01004832 JAG38772.1 GDHC01019310 JAP99318.1 GBHO01027564 JAG16040.1 DF973586 GAU35304.1 GBHO01002761 JAG40843.1 ABLF02002130 ABLF02010475 ABLF02010476 ABLF02041792 ABLF02003343 ABLF02041793 LJIG01022881 KRT78250.1 FJ544851 ACL97383.1 ABLF02020302 FJ544856 ACL97386.1 GECZ01019212 JAS50557.1 FJ544854 ACL97385.1 QGNW01001539 RVW37461.1 FJ544853 ACL97384.1 CM003611 KYP60991.1 GEZM01072005 JAV65635.1 KQ971969 KYB24675.1 QGNW01001128 RVW54433.1 QGNW01000024 RVX13462.1 GGMR01018348 MBY30967.1 AC079604 AC079131 AAG50698.1 AAG50765.1 AM464898 CAN74767.1 AM468226 CAN68235.1 AM466849 CAN77122.1 CM003608 KYP67178.1 QGNW01002171 RVW24539.1 QGNW01000748 RVW63137.1 FJ544857 ACL97387.1 QGNW01001661 RVW33963.1 AM451478 CAN80930.1 GBHO01028902 JAG14702.1 GBBI01004033 JAC14679.1 LBMM01010924 KMQ87150.1 QGNW01002379 RVW20302.1 NEVH01013252 PNF29308.1 DF974128 GAU45509.1 JXUM01031868 KQ560938 KXJ80258.1 QGNW01001434 RVW41591.1 GEZM01028916 JAV86195.1 PKPP01005057 PWA61635.1 DF974545 GAU49301.1 JAV86196.1 OIVN01004423 SPD17240.1 GEDC01029908 JAS07390.1 OIVN01004957 SPD20159.1 OIVN01002812 SPD06621.1 OIVN01001022 SPC88819.1 OIVN01000958 SPC87933.1 OIVN01004420 SPD17230.1 GDHC01020388 JAP98240.1 OIVN01000823 SPC86071.1 OIVN01001850 SPC98251.1 OIVN01003423 SPD11099.1 OIVN01006335 SPD30828.1 OIVN01004307 SPD16591.1 PKPP01006180 PWA57377.1 ASHM01016673 PNX98468.1 OIVN01000043 SPC73144.1 OIVN01001514 SPC94830.1 OIVN01005059 SPD20680.1 DS567821 PSQE01000003 RHN67289.1 QGNW01000165 RVW89346.1 AM425694 CAN62421.1 AM442619 CAN74029.1 LBMM01012772 KMQ86003.1 DF973357 GAU27529.1 OIVN01005261 SPD21666.1 OIVN01004456 SPD17553.1 AM453163 CAN69956.1 QZWG01000017 RZB57470.1 OIVN01005657 SPD23465.1 OIVN01002925 SPD07540.1 OIVN01006160 SPD26619.1 OIVN01005591 SPD23220.1 LVZM01023578 OUC39908.1
GBHO01004832 JAG38772.1 GDHC01019310 JAP99318.1 GBHO01027564 JAG16040.1 DF973586 GAU35304.1 GBHO01002761 JAG40843.1 ABLF02002130 ABLF02010475 ABLF02010476 ABLF02041792 ABLF02003343 ABLF02041793 LJIG01022881 KRT78250.1 FJ544851 ACL97383.1 ABLF02020302 FJ544856 ACL97386.1 GECZ01019212 JAS50557.1 FJ544854 ACL97385.1 QGNW01001539 RVW37461.1 FJ544853 ACL97384.1 CM003611 KYP60991.1 GEZM01072005 JAV65635.1 KQ971969 KYB24675.1 QGNW01001128 RVW54433.1 QGNW01000024 RVX13462.1 GGMR01018348 MBY30967.1 AC079604 AC079131 AAG50698.1 AAG50765.1 AM464898 CAN74767.1 AM468226 CAN68235.1 AM466849 CAN77122.1 CM003608 KYP67178.1 QGNW01002171 RVW24539.1 QGNW01000748 RVW63137.1 FJ544857 ACL97387.1 QGNW01001661 RVW33963.1 AM451478 CAN80930.1 GBHO01028902 JAG14702.1 GBBI01004033 JAC14679.1 LBMM01010924 KMQ87150.1 QGNW01002379 RVW20302.1 NEVH01013252 PNF29308.1 DF974128 GAU45509.1 JXUM01031868 KQ560938 KXJ80258.1 QGNW01001434 RVW41591.1 GEZM01028916 JAV86195.1 PKPP01005057 PWA61635.1 DF974545 GAU49301.1 JAV86196.1 OIVN01004423 SPD17240.1 GEDC01029908 JAS07390.1 OIVN01004957 SPD20159.1 OIVN01002812 SPD06621.1 OIVN01001022 SPC88819.1 OIVN01000958 SPC87933.1 OIVN01004420 SPD17230.1 GDHC01020388 JAP98240.1 OIVN01000823 SPC86071.1 OIVN01001850 SPC98251.1 OIVN01003423 SPD11099.1 OIVN01006335 SPD30828.1 OIVN01004307 SPD16591.1 PKPP01006180 PWA57377.1 ASHM01016673 PNX98468.1 OIVN01000043 SPC73144.1 OIVN01001514 SPC94830.1 OIVN01005059 SPD20680.1 DS567821 PSQE01000003 RHN67289.1 QGNW01000165 RVW89346.1 AM425694 CAN62421.1 AM442619 CAN74029.1 LBMM01012772 KMQ86003.1 DF973357 GAU27529.1 OIVN01005261 SPD21666.1 OIVN01004456 SPD17553.1 AM453163 CAN69956.1 QZWG01000017 RZB57470.1 OIVN01005657 SPD23465.1 OIVN01002925 SPD07540.1 OIVN01006160 SPD26619.1 OIVN01005591 SPD23220.1 LVZM01023578 OUC39908.1
Proteomes
Pfam
PF13976 gag_pre-integrs
+ More
PF07727 RVT_2
PF00665 rve
PF13961 DUF4219
PF00098 zf-CCHC
PF00931 NB-ARC
PF13855 LRR_8
PF14529 Exo_endo_phos_2
PF00078 RVT_1
PF17921 Integrase_H2C2
PF16770 RTT107_BRCT_5
PF00069 Pkinase
PF03171 2OG-FeII_Oxy
PF03936 Terpene_synth_C
PF01397 Terpene_synth
PF08263 LRRNT_2
PF00560 LRR_1
PF00005 ABC_tran
PF01061 ABC2_membrane
PF00182 Glyco_hydro_19
PF07727 RVT_2
PF00665 rve
PF13961 DUF4219
PF00098 zf-CCHC
PF00931 NB-ARC
PF13855 LRR_8
PF14529 Exo_endo_phos_2
PF00078 RVT_1
PF17921 Integrase_H2C2
PF16770 RTT107_BRCT_5
PF00069 Pkinase
PF03171 2OG-FeII_Oxy
PF03936 Terpene_synth_C
PF01397 Terpene_synth
PF08263 LRRNT_2
PF00560 LRR_1
PF00005 ABC_tran
PF01061 ABC2_membrane
PF00182 Glyco_hydro_19
Interpro
IPR001584
Integrase_cat-core
+ More
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR025724 GAG-pre-integrase_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR025314 DUF4219
IPR002182 NB-ARC
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR041588 Integrase_H2C2
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR027443 IPNS-like
IPR036965 Terpene_synth_N_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR034741 Terpene_cyclase-like_1_C
IPR001906 Terpene_synth_N
IPR013210 LRR_N_plant-typ
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR013525 ABC_2_trans
IPR023346 Lysozyme-like_dom_sf
IPR000726 Glyco_hydro_19_cat
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR025724 GAG-pre-integrase_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR025314 DUF4219
IPR002182 NB-ARC
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR041588 Integrase_H2C2
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR027443 IPNS-like
IPR036965 Terpene_synth_N_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR034741 Terpene_cyclase-like_1_C
IPR001906 Terpene_synth_N
IPR013210 LRR_N_plant-typ
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR013525 ABC_2_trans
IPR023346 Lysozyme-like_dom_sf
IPR000726 Glyco_hydro_19_cat
SUPFAM
Gene 3D
ProteinModelPortal
A0A0A9YSA9
A0A0A9YQT1
A0A0A9WQU6
A0A0A9Z0I6
A0A146KTS1
A0A0A9XG27
+ More
A0A2Z6MRL3 A0A0A9ZBW6 J9M5U1 J9LXN4 A0A0T6AT79 B8YLY3 X1WUQ9 B8YLY6 A0A1B6FK59 B8YLY5 A0A438DPU0 B8YLY4 A0A151T1X7 A0A1Y1L115 A0A139W9T1 A0A438F3A5 A0A438JWX0 A0A2S2PNK0 Q9C536 A5BMR0 A5BR97 A5BPR9 A0A151TJG4 A0A438CN15 A0A438FT52 B8YLY7 A0A438DEP9 A5B9M8 A0A0A9X1V9 A0A023F102 A0A0J7N374 A0A438CAN0 A0A2J7QL79 A0A2Z6PI76 A0A1U8HMK8 A0A182HBD9 A0A438E1L6 A0A1Y1MP14 A0A2U1MK50 A0A2Z6NYP5 A0A1Y1MQF0 A0A2N9HZS3 A0A1B6C2D3 A0A2N9I807 A0A2N9H4R5 A0A2N9FPD9 A0A2N9FL83 A0A2N9HZR4 A0A146KP10 A0A2N9FGL6 A0A2N9GG47 A0A1U8JW41 A0A2N9HHJ9 A0A2N9J203 A0A2N9HY48 A0A2U1M839 A0A2K3N600 A0A2N9EER7 A0A2N9G604 A0A2N9I9F7 H3H993 A0A396INR0 A0A438HY27 A5AFU8 A5B0R8 A0A0J7K689 A0A2Z6N7E5 A0A2N9I6M8 A0A2N9I0K6 A5BBA3 A0A445G8E4 A0A2N9IGN4 A0A2N9H7C1 A0A2N9IR42 A0A2N9IGI9 A0A1Y3E9R1
A0A2Z6MRL3 A0A0A9ZBW6 J9M5U1 J9LXN4 A0A0T6AT79 B8YLY3 X1WUQ9 B8YLY6 A0A1B6FK59 B8YLY5 A0A438DPU0 B8YLY4 A0A151T1X7 A0A1Y1L115 A0A139W9T1 A0A438F3A5 A0A438JWX0 A0A2S2PNK0 Q9C536 A5BMR0 A5BR97 A5BPR9 A0A151TJG4 A0A438CN15 A0A438FT52 B8YLY7 A0A438DEP9 A5B9M8 A0A0A9X1V9 A0A023F102 A0A0J7N374 A0A438CAN0 A0A2J7QL79 A0A2Z6PI76 A0A1U8HMK8 A0A182HBD9 A0A438E1L6 A0A1Y1MP14 A0A2U1MK50 A0A2Z6NYP5 A0A1Y1MQF0 A0A2N9HZS3 A0A1B6C2D3 A0A2N9I807 A0A2N9H4R5 A0A2N9FPD9 A0A2N9FL83 A0A2N9HZR4 A0A146KP10 A0A2N9FGL6 A0A2N9GG47 A0A1U8JW41 A0A2N9HHJ9 A0A2N9J203 A0A2N9HY48 A0A2U1M839 A0A2K3N600 A0A2N9EER7 A0A2N9G604 A0A2N9I9F7 H3H993 A0A396INR0 A0A438HY27 A5AFU8 A5B0R8 A0A0J7K689 A0A2Z6N7E5 A0A2N9I6M8 A0A2N9I0K6 A5BBA3 A0A445G8E4 A0A2N9IGN4 A0A2N9H7C1 A0A2N9IR42 A0A2N9IGI9 A0A1Y3E9R1
Ontologies
KEGG
GO
PANTHER
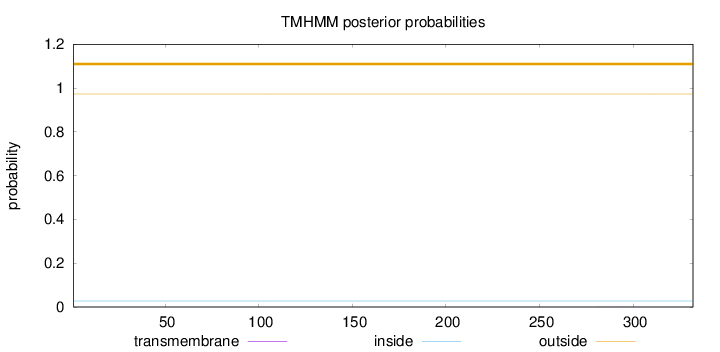
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0048
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02720
outside
1 - 332
Population Genetic Test Statistics
Pi
193.35394
Theta
195.565513
Tajima's D
0.148106
CLR
0
CSRT
0.409179541022949
Interpretation
Uncertain
