Gene
KWMTBOMO09982
Pre Gene Modal
BGIBMGA013202
Annotation
PREDICTED:_BRCA1-associated_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.64 Nuclear Reliability : 2.194
Sequence
CDS
ATGGCCGTCTCTTTGTGTGTACTACGTATTGAAATAGACGAAGAACTGTCCGAAATGGCAGACGGTGTTAACAATGAGGCCTCTAAGGACCAGAAAAGAAAGGACCGAGGAGCAAGGGAATCCAAAACCATAACAGTTGAAACATATGCTAGTGTTCTTAGTGGACCCAACACTGACCCAGGCATACTACCTACAAGTTCCCGCCGCTCACCTCCAGAGTCCAGCAGTATATCTGAAGACGATAACACTATTAGTTTTTTCTGTGGAAATCCACTAGTTGAAATTACTAAGGGCATATTACACTTGTACAAGGAAAATGAAATGAAAGAAGCAGAAGAAGCCAAGACACTTTGCCTACTCGCAGTTCCTGGTGCTTTAGGTGCACCCGACCTGCTTGCATTTGCTGCAGCTTGTCAACAGGATATTGCACATGTGAGGGTTCTCAGAGATGGTTCACCTGATCATTACATGGCACTATTAACATTTCGCACTAGTCAAGCAGCTCGAGAATTCCATGGGGCTTTTGATGGGGTTCCATATAGTAGTTTGGAACCAAATGCACTGTGTCATACAGCCTGGGTGTCAAGGGTTGAATGGGCAAAGGATGGTTCCCCACCACCATCACACACTGAGCTCCCTACTTGTCCTGTTTGTTTGGAGCGCATGGACGAGAGCGTGGCGGGCGTGCTGAGCGTGCAGTGCGCGCACGCCTTCCACGCGGAGTGTCTGGCGCGCAGCCGCGACCTGCGCTGCCCCGTGTGCCGCTGCGCGCAGACCCCCGAGCCGAGGGCGAGCGCACGCTGCCAGCTCTGCGACGCGCCCCACCGAGACGAGGAAGGCGACGACACCCGCGAGAACCTCAGCGGAGTCGCCGAGGGGCTCGTGGACGGGCTGGAGGAGGGCACGCTCTGGATATGCCTCATCTGTGGACACGTGGGATGTGGGAGGTACGAAGGGGGCCACGCCGCGAAGCACTTCCTGCAGTCCAACCACACGTACGCGCTGCAGCTCGGCTCCAACAGGGTCTGGGACTACGCAGGAGACAACTTCGTACACCGACTGGTGCAGAACAAGTCTGACGGCAAGCTGGTGGCGTCCGAGGGCGCCTGCGGGGGCAAGCACGACCACGAGTTCACCTACATACTCACCGAGCAACTGGACACGCAGCGCACTCACTACGAGGAGAAAATCGCCAAGCTGGAGGGGGAGCGGGCGGGGCTGGAGGGGGAGCGGGAGGCGGAGCGGGCGGCGCTGCAGGGGCGGGCGCGCGCCGCCGAGGCCCGGGCCGCTGACCTCGCCGCGCAGGTCGACGACCTCACGGCCCGCAACAAACAACTCGACCGGAAACTGCAGCAGCTCACCAACAAATTATCGGTTCTCCAAAACGAGTTGACGGAAGAGAGGGGCTTGGCGGCGGCCATGGCCAGCAGCCAGTCGGAGTGGCAGGCCAGGGCTAAAGAAAAGGAGGAGAATTTCACCAAAGAGGTCAGCGATCTGAAGGAGCAACTCCGCGACGTGATGTTCTTTGTGGAGGCGCGAGCGCAGCTGGAGCGAAGACTATGTACTTGA
Protein
MAVSLCVLRIEIDEELSEMADGVNNEASKDQKRKDRGARESKTITVETYASVLSGPNTDPGILPTSSRRSPPESSSISEDDNTISFFCGNPLVEITKGILHLYKENEMKEAEEAKTLCLLAVPGALGAPDLLAFAAACQQDIAHVRVLRDGSPDHYMALLTFRTSQAAREFHGAFDGVPYSSLEPNALCHTAWVSRVEWAKDGSPPPSHTELPTCPVCLERMDESVAGVLSVQCAHAFHAECLARSRDLRCPVCRCAQTPEPRASARCQLCDAPHRDEEGDDTRENLSGVAEGLVDGLEEGTLWICLICGHVGCGRYEGGHAAKHFLQSNHTYALQLGSNRVWDYAGDNFVHRLVQNKSDGKLVASEGACGGKHDHEFTYILTEQLDTQRTHYEEKIAKLEGERAGLEGEREAERAALQGRARAAEARAADLAAQVDDLTARNKQLDRKLQQLTNKLSVLQNELTEERGLAAAMASSQSEWQARAKEKEENFTKEVSDLKEQLRDVMFFVEARAQLERRLCT
Summary
Uniprot
A0A2A4JH71
A0A437BLM8
A0A194PRP4
S4PXK4
A0A1E1W540
A0A1E1WCY9
+ More
A0A0N1PHU3 D2A5C1 A0A1Y1LUA4 U5ESP9 A0A1L8DTH2 A0A1B0CVF8 Q177G7 A0A182G3F9 A0A1J1HS73 A0A084VJA9 B4LZK5 A0A0Q9WR96 A0A1Q3FBJ2 B0W941 A0A182IZD4 B4KD18 A0A0M4EW95 A0A151JZW2 A0A1B6H9S7 B3P363 A0A336LY22 A0A2M3Z681 W5JHW4 A0A067RBB1 A0A195AZ85 A0A2M3Z5C3 E2A8Y9 A0A0J7KNL5 F4W4B5 B4PMS7 A0A158NMI5 A0A182FRB0 A0A2M4BHH2 A0A2M4AG93 A0A1I8NZ06 A0A2M4AGB4 B4JUX3 B4NIT3 A0A182PNE0 A0A151XH94 A0A3B0JNT2 A0A026WFF1 A0A3L8DJK2 A0A182MAH7 N6U3R6 E2BA11 A0A3B0JNH6 A0A182Q5G0 Q9VDZ1 A0A0B4KHR2 A0A182Y466 A0A1B6L8D8 B3M2F3 B4IHX9 A0A1W4UN00 A0A182W023 Q293Q0 B4GLT1 A0A151IY98 A0A2J7PX41 A0A1B6F5P3 A0A182XGJ1 A0A182HZT5 A0A182VFT5 A0A182LQW2 F5HMZ5 A0A0L0BQZ1 A0A182UBF9 A0A1A9Y6P5 A0A1B0BNK6 A0A2J7PX57 Q7PYV7 A0A088ATA2 A0A310S763 A0A2A3EI35 A0A182RNV7 A0A195CX58 A0A1B0G7Z4 A0A154NWZ6 A0A1A9VB81 T1PGB2 A0A1I8N9H3 A0A0M9AB45 A0A182N682 A0A182JUI4 A0A232F1X9 K7IYZ0 A0A0K8UZ32 A0A034W5L1 E9IXK5 A0A0A1X4T7
A0A0N1PHU3 D2A5C1 A0A1Y1LUA4 U5ESP9 A0A1L8DTH2 A0A1B0CVF8 Q177G7 A0A182G3F9 A0A1J1HS73 A0A084VJA9 B4LZK5 A0A0Q9WR96 A0A1Q3FBJ2 B0W941 A0A182IZD4 B4KD18 A0A0M4EW95 A0A151JZW2 A0A1B6H9S7 B3P363 A0A336LY22 A0A2M3Z681 W5JHW4 A0A067RBB1 A0A195AZ85 A0A2M3Z5C3 E2A8Y9 A0A0J7KNL5 F4W4B5 B4PMS7 A0A158NMI5 A0A182FRB0 A0A2M4BHH2 A0A2M4AG93 A0A1I8NZ06 A0A2M4AGB4 B4JUX3 B4NIT3 A0A182PNE0 A0A151XH94 A0A3B0JNT2 A0A026WFF1 A0A3L8DJK2 A0A182MAH7 N6U3R6 E2BA11 A0A3B0JNH6 A0A182Q5G0 Q9VDZ1 A0A0B4KHR2 A0A182Y466 A0A1B6L8D8 B3M2F3 B4IHX9 A0A1W4UN00 A0A182W023 Q293Q0 B4GLT1 A0A151IY98 A0A2J7PX41 A0A1B6F5P3 A0A182XGJ1 A0A182HZT5 A0A182VFT5 A0A182LQW2 F5HMZ5 A0A0L0BQZ1 A0A182UBF9 A0A1A9Y6P5 A0A1B0BNK6 A0A2J7PX57 Q7PYV7 A0A088ATA2 A0A310S763 A0A2A3EI35 A0A182RNV7 A0A195CX58 A0A1B0G7Z4 A0A154NWZ6 A0A1A9VB81 T1PGB2 A0A1I8N9H3 A0A0M9AB45 A0A182N682 A0A182JUI4 A0A232F1X9 K7IYZ0 A0A0K8UZ32 A0A034W5L1 E9IXK5 A0A0A1X4T7
Pubmed
26354079
23622113
18362917
19820115
28004739
17510324
+ More
26483478 24438588 17994087 18057021 20920257 23761445 24845553 20798317 21719571 17550304 21347285 24508170 30249741 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 15632085 20966253 12364791 14747013 17210077 26108605 25315136 28648823 20075255 25348373 21282665 25830018
26483478 24438588 17994087 18057021 20920257 23761445 24845553 20798317 21719571 17550304 21347285 24508170 30249741 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 15632085 20966253 12364791 14747013 17210077 26108605 25315136 28648823 20075255 25348373 21282665 25830018
EMBL
NWSH01001412
PCG71321.1
RSAL01000036
RVE51241.1
KQ459595
KPI95982.1
+ More
GAIX01004883 JAA87677.1 GDQN01008957 JAT82097.1 GDQN01006192 JAT84862.1 KQ460643 KPJ13314.1 KQ971345 EFA05346.2 GEZM01046441 JAV77232.1 GANO01003164 JAB56707.1 GFDF01004373 JAV09711.1 AJWK01030488 CH477376 EAT42310.1 JXUM01040801 KQ561259 KXJ79188.1 CVRI01000020 CRK90903.1 ATLV01013500 KE524866 KFB38053.1 CH940650 EDW67144.2 KRF83132.1 KRF83131.1 GFDL01010104 JAV24941.1 DS231862 EDS39738.1 CH933806 EDW13788.1 CP012526 ALC47767.1 KQ981360 KYN42443.1 GECU01036312 JAS71394.1 CH954181 EDV48515.1 UFQT01000197 SSX21573.1 GGFM01003270 MBW24021.1 ADMH02001204 ETN63711.1 KK852619 KDR20118.1 KQ976697 KYM77516.1 GGFM01002962 MBW23713.1 GL437703 EFN70088.1 LBMM01005070 KMQ91841.1 GL887503 EGI70959.1 CM000160 EDW95997.2 ADTU01020596 ADTU01020597 GGFJ01003369 MBW52510.1 GGFK01006489 MBW39810.1 GGFK01006503 MBW39824.1 CH916374 EDV91293.1 CH964272 EDW83797.1 KQ982149 KYQ59580.1 OUUW01000008 SPP83785.1 KK107238 EZA54648.1 QOIP01000007 RLU20614.1 AXCM01000193 APGK01050705 KB741175 ENN73217.1 GL446638 EFN87466.1 SPP83784.1 AXCN02000271 AE014297 AY047557 AAF55646.2 AAK77289.1 AGB96104.1 GEBQ01020183 JAT19794.1 CH902617 EDV43406.2 CH480840 EDW49496.1 CM000070 EAL29164.2 CH479185 EDW38505.1 KQ980790 KYN13154.1 NEVH01020862 PNF20905.1 GECZ01024576 JAS45193.1 APCN01002010 AAAB01008987 EGK97376.1 JRES01001498 KNC22452.1 JXJN01017426 PNF20904.1 EAA00940.4 KQ766607 OAD53617.1 KZ288253 PBC30932.1 KQ977231 KYN04744.1 CCAG010010583 KQ434777 KZC04189.1 KA647847 AFP62476.1 KQ435694 KOX80917.1 NNAY01001189 OXU24806.1 GDHF01020463 JAI31851.1 GAKP01009375 JAC49577.1 GL766755 EFZ14699.1 GBXI01007988 JAD06304.1
GAIX01004883 JAA87677.1 GDQN01008957 JAT82097.1 GDQN01006192 JAT84862.1 KQ460643 KPJ13314.1 KQ971345 EFA05346.2 GEZM01046441 JAV77232.1 GANO01003164 JAB56707.1 GFDF01004373 JAV09711.1 AJWK01030488 CH477376 EAT42310.1 JXUM01040801 KQ561259 KXJ79188.1 CVRI01000020 CRK90903.1 ATLV01013500 KE524866 KFB38053.1 CH940650 EDW67144.2 KRF83132.1 KRF83131.1 GFDL01010104 JAV24941.1 DS231862 EDS39738.1 CH933806 EDW13788.1 CP012526 ALC47767.1 KQ981360 KYN42443.1 GECU01036312 JAS71394.1 CH954181 EDV48515.1 UFQT01000197 SSX21573.1 GGFM01003270 MBW24021.1 ADMH02001204 ETN63711.1 KK852619 KDR20118.1 KQ976697 KYM77516.1 GGFM01002962 MBW23713.1 GL437703 EFN70088.1 LBMM01005070 KMQ91841.1 GL887503 EGI70959.1 CM000160 EDW95997.2 ADTU01020596 ADTU01020597 GGFJ01003369 MBW52510.1 GGFK01006489 MBW39810.1 GGFK01006503 MBW39824.1 CH916374 EDV91293.1 CH964272 EDW83797.1 KQ982149 KYQ59580.1 OUUW01000008 SPP83785.1 KK107238 EZA54648.1 QOIP01000007 RLU20614.1 AXCM01000193 APGK01050705 KB741175 ENN73217.1 GL446638 EFN87466.1 SPP83784.1 AXCN02000271 AE014297 AY047557 AAF55646.2 AAK77289.1 AGB96104.1 GEBQ01020183 JAT19794.1 CH902617 EDV43406.2 CH480840 EDW49496.1 CM000070 EAL29164.2 CH479185 EDW38505.1 KQ980790 KYN13154.1 NEVH01020862 PNF20905.1 GECZ01024576 JAS45193.1 APCN01002010 AAAB01008987 EGK97376.1 JRES01001498 KNC22452.1 JXJN01017426 PNF20904.1 EAA00940.4 KQ766607 OAD53617.1 KZ288253 PBC30932.1 KQ977231 KYN04744.1 CCAG010010583 KQ434777 KZC04189.1 KA647847 AFP62476.1 KQ435694 KOX80917.1 NNAY01001189 OXU24806.1 GDHF01020463 JAI31851.1 GAKP01009375 JAC49577.1 GL766755 EFZ14699.1 GBXI01007988 JAD06304.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007266
UP000092461
+ More
UP000008820 UP000069940 UP000249989 UP000183832 UP000030765 UP000008792 UP000002320 UP000075880 UP000009192 UP000092553 UP000078541 UP000008711 UP000000673 UP000027135 UP000078540 UP000000311 UP000036403 UP000007755 UP000002282 UP000005205 UP000069272 UP000095300 UP000001070 UP000007798 UP000075885 UP000075809 UP000268350 UP000053097 UP000279307 UP000075883 UP000019118 UP000008237 UP000075886 UP000000803 UP000076408 UP000007801 UP000001292 UP000192221 UP000075920 UP000001819 UP000008744 UP000078492 UP000235965 UP000076407 UP000075840 UP000075903 UP000075882 UP000007062 UP000037069 UP000075902 UP000092443 UP000092460 UP000005203 UP000242457 UP000075900 UP000078542 UP000092444 UP000076502 UP000078200 UP000095301 UP000053105 UP000075884 UP000075881 UP000215335 UP000002358
UP000008820 UP000069940 UP000249989 UP000183832 UP000030765 UP000008792 UP000002320 UP000075880 UP000009192 UP000092553 UP000078541 UP000008711 UP000000673 UP000027135 UP000078540 UP000000311 UP000036403 UP000007755 UP000002282 UP000005205 UP000069272 UP000095300 UP000001070 UP000007798 UP000075885 UP000075809 UP000268350 UP000053097 UP000279307 UP000075883 UP000019118 UP000008237 UP000075886 UP000000803 UP000076408 UP000007801 UP000001292 UP000192221 UP000075920 UP000001819 UP000008744 UP000078492 UP000235965 UP000076407 UP000075840 UP000075903 UP000075882 UP000007062 UP000037069 UP000075902 UP000092443 UP000092460 UP000005203 UP000242457 UP000075900 UP000078542 UP000092444 UP000076502 UP000078200 UP000095301 UP000053105 UP000075884 UP000075881 UP000215335 UP000002358
PRIDE
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
A0A2A4JH71
A0A437BLM8
A0A194PRP4
S4PXK4
A0A1E1W540
A0A1E1WCY9
+ More
A0A0N1PHU3 D2A5C1 A0A1Y1LUA4 U5ESP9 A0A1L8DTH2 A0A1B0CVF8 Q177G7 A0A182G3F9 A0A1J1HS73 A0A084VJA9 B4LZK5 A0A0Q9WR96 A0A1Q3FBJ2 B0W941 A0A182IZD4 B4KD18 A0A0M4EW95 A0A151JZW2 A0A1B6H9S7 B3P363 A0A336LY22 A0A2M3Z681 W5JHW4 A0A067RBB1 A0A195AZ85 A0A2M3Z5C3 E2A8Y9 A0A0J7KNL5 F4W4B5 B4PMS7 A0A158NMI5 A0A182FRB0 A0A2M4BHH2 A0A2M4AG93 A0A1I8NZ06 A0A2M4AGB4 B4JUX3 B4NIT3 A0A182PNE0 A0A151XH94 A0A3B0JNT2 A0A026WFF1 A0A3L8DJK2 A0A182MAH7 N6U3R6 E2BA11 A0A3B0JNH6 A0A182Q5G0 Q9VDZ1 A0A0B4KHR2 A0A182Y466 A0A1B6L8D8 B3M2F3 B4IHX9 A0A1W4UN00 A0A182W023 Q293Q0 B4GLT1 A0A151IY98 A0A2J7PX41 A0A1B6F5P3 A0A182XGJ1 A0A182HZT5 A0A182VFT5 A0A182LQW2 F5HMZ5 A0A0L0BQZ1 A0A182UBF9 A0A1A9Y6P5 A0A1B0BNK6 A0A2J7PX57 Q7PYV7 A0A088ATA2 A0A310S763 A0A2A3EI35 A0A182RNV7 A0A195CX58 A0A1B0G7Z4 A0A154NWZ6 A0A1A9VB81 T1PGB2 A0A1I8N9H3 A0A0M9AB45 A0A182N682 A0A182JUI4 A0A232F1X9 K7IYZ0 A0A0K8UZ32 A0A034W5L1 E9IXK5 A0A0A1X4T7
A0A0N1PHU3 D2A5C1 A0A1Y1LUA4 U5ESP9 A0A1L8DTH2 A0A1B0CVF8 Q177G7 A0A182G3F9 A0A1J1HS73 A0A084VJA9 B4LZK5 A0A0Q9WR96 A0A1Q3FBJ2 B0W941 A0A182IZD4 B4KD18 A0A0M4EW95 A0A151JZW2 A0A1B6H9S7 B3P363 A0A336LY22 A0A2M3Z681 W5JHW4 A0A067RBB1 A0A195AZ85 A0A2M3Z5C3 E2A8Y9 A0A0J7KNL5 F4W4B5 B4PMS7 A0A158NMI5 A0A182FRB0 A0A2M4BHH2 A0A2M4AG93 A0A1I8NZ06 A0A2M4AGB4 B4JUX3 B4NIT3 A0A182PNE0 A0A151XH94 A0A3B0JNT2 A0A026WFF1 A0A3L8DJK2 A0A182MAH7 N6U3R6 E2BA11 A0A3B0JNH6 A0A182Q5G0 Q9VDZ1 A0A0B4KHR2 A0A182Y466 A0A1B6L8D8 B3M2F3 B4IHX9 A0A1W4UN00 A0A182W023 Q293Q0 B4GLT1 A0A151IY98 A0A2J7PX41 A0A1B6F5P3 A0A182XGJ1 A0A182HZT5 A0A182VFT5 A0A182LQW2 F5HMZ5 A0A0L0BQZ1 A0A182UBF9 A0A1A9Y6P5 A0A1B0BNK6 A0A2J7PX57 Q7PYV7 A0A088ATA2 A0A310S763 A0A2A3EI35 A0A182RNV7 A0A195CX58 A0A1B0G7Z4 A0A154NWZ6 A0A1A9VB81 T1PGB2 A0A1I8N9H3 A0A0M9AB45 A0A182N682 A0A182JUI4 A0A232F1X9 K7IYZ0 A0A0K8UZ32 A0A034W5L1 E9IXK5 A0A0A1X4T7
PDB
5G0F
E-value=3.02741e-05,
Score=114
Ontologies
GO
Topology
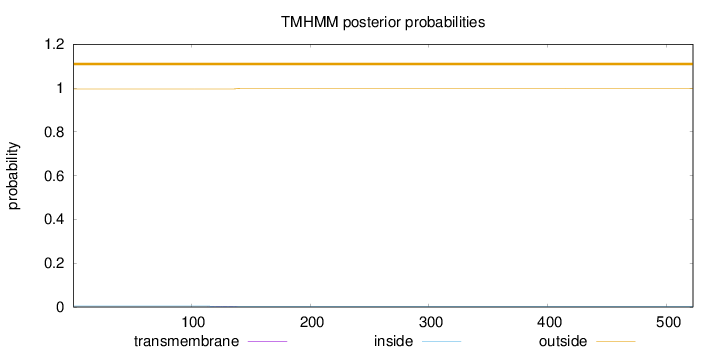
Length:
522
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0763300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00467
outside
1 - 522
Population Genetic Test Statistics
Pi
277.329663
Theta
186.861325
Tajima's D
1.624481
CLR
0.500547
CSRT
0.813009349532523
Interpretation
Uncertain
