Gene
KWMTBOMO09980 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013201
Annotation
14-3-3_epsilon_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.898
Sequence
CDS
ATGTCGGAAAGGGAAGATAATGTGTATAAAGCAAAGTTAGCTGAGCAGGCTGAGCGCTATGACGAAATGGTGGAGGCGATGAAGAATGTAGCATCACGCAACGTGTCCGACAATGAGCTGACAGTGGAGGAGAGGAACCTGCTGTCGGTCGCCTACAAGAACGTGATCGGCGCCCGCCGCGCCTCCTGGCGGATCATCTCCTCCATCGAACAGAAGGAAGAAACAAAAGGCGCAGAAGACAAATTGAACATGATCAGAGCGTACCGCAGCCAGGTTGAGAAGGAGCTCCGTGACATCTGCTCTGACATCCTCGGTGTCCTCGACAAGTACCTCATCCCCAGCTCACAGACCGGAGAATCAAAAGTTTTTTATTACAAAATGAAGGGCGACTACCACCGGTACCTGGCCGAGTTCGCGACCGGCAACGACCGCAAGGAGGCTGCCGAGAACTCGCTGGTCGCGTACAAGGCAGCCTCCGACATCGCCATGACCGAACTGCCCCCCACCCACCCCATCCGCCTCGGACTCGCGCTCAACTTCTCCGTGTTCTACTACGAGATCCTGAACAGCCCGGACCGCGCCTGCCGGCTCGCCAAGGCCGCCTTCGACGACGCCATCGCCGAACTGGACACGCTCTCGGAGGAGAGCTACAAGGACTCCACGCTCATCATGCAGCTGCTGCGGGACAACCTCACGCTGTGGACCTCCGACATGCAGGGCGACGGGGAGTCCGCCGACGCCGAGCAGAAGGAGCCGGCGCAGGACGGCGAGGACCAGGACGTCTCGTAA
Protein
MSEREDNVYKAKLAEQAERYDEMVEAMKNVASRNVSDNELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEETKGAEDKLNMIRAYRSQVEKELRDICSDILGVLDKYLIPSSQTGESKVFYYKMKGDYHRYLAEFATGNDRKEAAENSLVAYKAASDIAMTELPPTHPIRLGLALNFSVFYYEILNSPDRACRLAKAAFDDAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDMQGDGESADAEQKEPAQDGEDQDVS
Summary
Similarity
Belongs to the 14-3-3 family.
Uniprot
Q1HPT4
H9JUI8
A0A1E1WG99
S4PAI5
A0A0N1I8S8
A0A2H1VA22
+ More
A0A194PXV4 D9IX59 A0A3S2NM79 A0A1V1FQH9 A0A191UR67 G9C5F4 F4WPA6 A0A158NR28 E2ADN2 E9IT60 A0A2J7PX37 A0A1B6GNF1 A0A1B6D1Q1 A0A158NR27 A0A1B6I1T4 A0A3L8DWU3 R4WP03 K7J834 E2B527 A0A0T6B240 U5EZA7 E9H8M4 A0A0P4YC34 A0A1W4X1P4 A0A1Y1K6X2 A0A0M4MP46 A0A0D3RLG3 D6WYT2 A0A0P5TTR4 A0A0A9WRP1 V9IEG2 A0A087ZUP0 A0A0P6HQU0 A0A336K4X4 J3JVF7 T1DIH4 A0A1Q3EU84 T1DP10 A0A2M3YYK6 A0A2M4A1R3 W5J8H0 A0A1Y1KBX6 A0A084W0J2 A0A182IR93 A0A0K8TF46 T1II32 A0A087TZB3 A0A0P5VWS6 U4ULQ8 A0A2P8ZJQ3 A0A0K8TPL0 A0A0P6IIL4 A0A182XP04 A0A182YND2 A0A182VMX6 Q7PX08 A0A182HK46 A0A131Y0H7 B7Q5I4 A0A182Q9Y3 A0A0P5YN02 A0A224XIF0 A0A0V0G7I4 A0A023F9I8 A0A069DQV0 A0A023ENU3 A0A182MR44 A0A0P4VWN9 R4G8P2 A0A182PR85 A0A1I8NMW4 A0A0L0BZI1 A0A1L8EHB0 T1PM25 A0A1Z5L731 A0A224YU45 A0A131YYE0 L7M840 A0A131XIE5 A0A1L8DU98 A0A1A9YNT0 A0A1A9UCL9 A0A1B0BFC3 A0A1B0AIP9 A0A1A9X228 D3TRP1 A0A411G7P4 A0A2R5L8P8 A0A293MSY7 A0A385L4G2 A0A195E6J1 Q16QZ7 A0A195EU94 W8C765
A0A194PXV4 D9IX59 A0A3S2NM79 A0A1V1FQH9 A0A191UR67 G9C5F4 F4WPA6 A0A158NR28 E2ADN2 E9IT60 A0A2J7PX37 A0A1B6GNF1 A0A1B6D1Q1 A0A158NR27 A0A1B6I1T4 A0A3L8DWU3 R4WP03 K7J834 E2B527 A0A0T6B240 U5EZA7 E9H8M4 A0A0P4YC34 A0A1W4X1P4 A0A1Y1K6X2 A0A0M4MP46 A0A0D3RLG3 D6WYT2 A0A0P5TTR4 A0A0A9WRP1 V9IEG2 A0A087ZUP0 A0A0P6HQU0 A0A336K4X4 J3JVF7 T1DIH4 A0A1Q3EU84 T1DP10 A0A2M3YYK6 A0A2M4A1R3 W5J8H0 A0A1Y1KBX6 A0A084W0J2 A0A182IR93 A0A0K8TF46 T1II32 A0A087TZB3 A0A0P5VWS6 U4ULQ8 A0A2P8ZJQ3 A0A0K8TPL0 A0A0P6IIL4 A0A182XP04 A0A182YND2 A0A182VMX6 Q7PX08 A0A182HK46 A0A131Y0H7 B7Q5I4 A0A182Q9Y3 A0A0P5YN02 A0A224XIF0 A0A0V0G7I4 A0A023F9I8 A0A069DQV0 A0A023ENU3 A0A182MR44 A0A0P4VWN9 R4G8P2 A0A182PR85 A0A1I8NMW4 A0A0L0BZI1 A0A1L8EHB0 T1PM25 A0A1Z5L731 A0A224YU45 A0A131YYE0 L7M840 A0A131XIE5 A0A1L8DU98 A0A1A9YNT0 A0A1A9UCL9 A0A1B0BFC3 A0A1B0AIP9 A0A1A9X228 D3TRP1 A0A411G7P4 A0A2R5L8P8 A0A293MSY7 A0A385L4G2 A0A195E6J1 Q16QZ7 A0A195EU94 W8C765
Pubmed
17949913
18462820
19121390
23622113
26354079
28410430
+ More
27142481 21835214 21719571 21347285 20798317 21282665 30249741 23691247 20075255 21292972 28004739 18362917 19820115 25401762 26823975 22516182 24330624 20920257 23761445 24438588 23537049 29403074 26369729 25244985 12364791 14747013 17210077 25474469 26334808 24945155 27129103 26108605 25315136 28528879 28797301 26830274 25576852 28049606 20353571 17510324 24495485
27142481 21835214 21719571 21347285 20798317 21282665 30249741 23691247 20075255 21292972 28004739 18362917 19820115 25401762 26823975 22516182 24330624 20920257 23761445 24438588 23537049 29403074 26369729 25244985 12364791 14747013 17210077 25474469 26334808 24945155 27129103 26108605 25315136 28528879 28797301 26830274 25576852 28049606 20353571 17510324 24495485
EMBL
DQ443318
EF112400
AB378098
ABF51407.1
ABK97426.1
BAG38534.1
+ More
BABH01022568 GDQN01005049 JAT86005.1 GAIX01003249 JAA89311.1 KQ460655 KPJ12926.1 ODYU01001198 SOQ37084.1 KQ459595 KPI95980.1 HM215623 ADK12634.1 RSAL01000036 RVE51244.1 FX985370 BAX07383.1 KU365952 ANJ04668.1 HQ851395 AEV89773.1 GL888243 EGI64087.1 ADTU01023768 ADTU01023769 GL438820 EFN68394.1 GL765434 EFZ16331.1 NEVH01020862 PNF20884.1 GECZ01005896 JAS63873.1 GEDC01017669 JAS19629.1 GECU01026814 JAS80892.1 QOIP01000003 RLU24940.1 AK417401 BAN20616.1 GL445685 EFN89235.1 LJIG01016250 KRT81166.1 GANO01001134 JAB58737.1 GL732605 EFX71844.1 GDIP01239200 GDIP01195898 GDIP01062161 LRGB01000084 JAI84201.1 KZS20952.1 GEZM01091070 GEZM01091068 JAV57124.1 KR075845 ALE20564.1 KP099937 AJS10721.1 KQ971357 EFA08461.1 GDIP01199295 GDIP01122099 JAL81615.1 GBHO01036064 GDHC01016643 JAG07540.1 JAQ01986.1 JR039817 AEY58856.1 GDIQ01023427 JAN71310.1 UFQS01000046 UFQT01000046 SSW98495.1 SSX18881.1 BT127225 AEE62187.1 GALA01000931 JAA93921.1 GFDL01016178 JAV18867.1 GAMD01002375 JAA99215.1 GGFM01000584 MBW21335.1 GGFK01001349 MBW34670.1 ADMH02002066 GGFL01004302 ETN59683.1 MBW68480.1 GEZM01091069 GEZM01091067 JAV57125.1 ATLV01019122 KE525262 KFB43736.1 AXCP01002654 GBRD01001604 JAG64217.1 JH430086 KK117428 KFM70452.1 GDIP01094156 JAM09559.1 KB632376 ERL94057.1 PYGN01000035 PSN56726.1 GDAI01001520 JAI16083.1 GDIQ01004626 JAN90111.1 AAAB01008987 EAA01035.2 APCN01000816 GEFM01003821 JAP71975.1 ABJB010962481 DS861388 EEC14106.1 AXCN02001485 GDIP01057361 JAM46354.1 GFTR01004169 JAW12257.1 GECL01002813 JAP03311.1 GBBI01000565 JAC18147.1 GBGD01002624 JAC86265.1 GAPW01003484 JAC10114.1 AXCM01000964 GDKW01001486 JAI55109.1 GAHY01000627 JAA76883.1 JRES01001197 KNC24654.1 GFDG01000741 JAV18058.1 KA644591 KA649779 AFP64408.1 GFJQ02003890 JAW03080.1 GFPF01009982 MAA21128.1 GEDV01004348 JAP84209.1 GACK01005760 JAA59274.1 GEFH01001692 JAP66889.1 GFDF01004075 JAV10009.1 JXJN01013364 CCAG010012184 EZ424093 ADD20369.1 MH366130 QBB01939.1 GGLE01001709 MBY05835.1 GFWV01019255 MAA43983.1 MH538272 AYA22648.1 KQ979608 KYN20492.1 CH477727 EAT36824.1 EAT36825.1 EJY57904.1 KQ981965 KYN31818.1 GAMC01000732 JAC05824.1
BABH01022568 GDQN01005049 JAT86005.1 GAIX01003249 JAA89311.1 KQ460655 KPJ12926.1 ODYU01001198 SOQ37084.1 KQ459595 KPI95980.1 HM215623 ADK12634.1 RSAL01000036 RVE51244.1 FX985370 BAX07383.1 KU365952 ANJ04668.1 HQ851395 AEV89773.1 GL888243 EGI64087.1 ADTU01023768 ADTU01023769 GL438820 EFN68394.1 GL765434 EFZ16331.1 NEVH01020862 PNF20884.1 GECZ01005896 JAS63873.1 GEDC01017669 JAS19629.1 GECU01026814 JAS80892.1 QOIP01000003 RLU24940.1 AK417401 BAN20616.1 GL445685 EFN89235.1 LJIG01016250 KRT81166.1 GANO01001134 JAB58737.1 GL732605 EFX71844.1 GDIP01239200 GDIP01195898 GDIP01062161 LRGB01000084 JAI84201.1 KZS20952.1 GEZM01091070 GEZM01091068 JAV57124.1 KR075845 ALE20564.1 KP099937 AJS10721.1 KQ971357 EFA08461.1 GDIP01199295 GDIP01122099 JAL81615.1 GBHO01036064 GDHC01016643 JAG07540.1 JAQ01986.1 JR039817 AEY58856.1 GDIQ01023427 JAN71310.1 UFQS01000046 UFQT01000046 SSW98495.1 SSX18881.1 BT127225 AEE62187.1 GALA01000931 JAA93921.1 GFDL01016178 JAV18867.1 GAMD01002375 JAA99215.1 GGFM01000584 MBW21335.1 GGFK01001349 MBW34670.1 ADMH02002066 GGFL01004302 ETN59683.1 MBW68480.1 GEZM01091069 GEZM01091067 JAV57125.1 ATLV01019122 KE525262 KFB43736.1 AXCP01002654 GBRD01001604 JAG64217.1 JH430086 KK117428 KFM70452.1 GDIP01094156 JAM09559.1 KB632376 ERL94057.1 PYGN01000035 PSN56726.1 GDAI01001520 JAI16083.1 GDIQ01004626 JAN90111.1 AAAB01008987 EAA01035.2 APCN01000816 GEFM01003821 JAP71975.1 ABJB010962481 DS861388 EEC14106.1 AXCN02001485 GDIP01057361 JAM46354.1 GFTR01004169 JAW12257.1 GECL01002813 JAP03311.1 GBBI01000565 JAC18147.1 GBGD01002624 JAC86265.1 GAPW01003484 JAC10114.1 AXCM01000964 GDKW01001486 JAI55109.1 GAHY01000627 JAA76883.1 JRES01001197 KNC24654.1 GFDG01000741 JAV18058.1 KA644591 KA649779 AFP64408.1 GFJQ02003890 JAW03080.1 GFPF01009982 MAA21128.1 GEDV01004348 JAP84209.1 GACK01005760 JAA59274.1 GEFH01001692 JAP66889.1 GFDF01004075 JAV10009.1 JXJN01013364 CCAG010012184 EZ424093 ADD20369.1 MH366130 QBB01939.1 GGLE01001709 MBY05835.1 GFWV01019255 MAA43983.1 MH538272 AYA22648.1 KQ979608 KYN20492.1 CH477727 EAT36824.1 EAT36825.1 EJY57904.1 KQ981965 KYN31818.1 GAMC01000732 JAC05824.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007755
UP000005205
+ More
UP000000311 UP000235965 UP000279307 UP000002358 UP000008237 UP000000305 UP000076858 UP000192223 UP000007266 UP000005203 UP000000673 UP000030765 UP000075880 UP000054359 UP000030742 UP000245037 UP000076407 UP000076408 UP000075903 UP000007062 UP000075840 UP000001555 UP000075886 UP000075883 UP000075885 UP000095300 UP000037069 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000078492 UP000008820 UP000078541
UP000000311 UP000235965 UP000279307 UP000002358 UP000008237 UP000000305 UP000076858 UP000192223 UP000007266 UP000005203 UP000000673 UP000030765 UP000075880 UP000054359 UP000030742 UP000245037 UP000076407 UP000076408 UP000075903 UP000007062 UP000075840 UP000001555 UP000075886 UP000075883 UP000075885 UP000095300 UP000037069 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000078492 UP000008820 UP000078541
Pfam
PF00244 14-3-3
Interpro
SUPFAM
SSF48445
SSF48445
Gene 3D
ProteinModelPortal
Q1HPT4
H9JUI8
A0A1E1WG99
S4PAI5
A0A0N1I8S8
A0A2H1VA22
+ More
A0A194PXV4 D9IX59 A0A3S2NM79 A0A1V1FQH9 A0A191UR67 G9C5F4 F4WPA6 A0A158NR28 E2ADN2 E9IT60 A0A2J7PX37 A0A1B6GNF1 A0A1B6D1Q1 A0A158NR27 A0A1B6I1T4 A0A3L8DWU3 R4WP03 K7J834 E2B527 A0A0T6B240 U5EZA7 E9H8M4 A0A0P4YC34 A0A1W4X1P4 A0A1Y1K6X2 A0A0M4MP46 A0A0D3RLG3 D6WYT2 A0A0P5TTR4 A0A0A9WRP1 V9IEG2 A0A087ZUP0 A0A0P6HQU0 A0A336K4X4 J3JVF7 T1DIH4 A0A1Q3EU84 T1DP10 A0A2M3YYK6 A0A2M4A1R3 W5J8H0 A0A1Y1KBX6 A0A084W0J2 A0A182IR93 A0A0K8TF46 T1II32 A0A087TZB3 A0A0P5VWS6 U4ULQ8 A0A2P8ZJQ3 A0A0K8TPL0 A0A0P6IIL4 A0A182XP04 A0A182YND2 A0A182VMX6 Q7PX08 A0A182HK46 A0A131Y0H7 B7Q5I4 A0A182Q9Y3 A0A0P5YN02 A0A224XIF0 A0A0V0G7I4 A0A023F9I8 A0A069DQV0 A0A023ENU3 A0A182MR44 A0A0P4VWN9 R4G8P2 A0A182PR85 A0A1I8NMW4 A0A0L0BZI1 A0A1L8EHB0 T1PM25 A0A1Z5L731 A0A224YU45 A0A131YYE0 L7M840 A0A131XIE5 A0A1L8DU98 A0A1A9YNT0 A0A1A9UCL9 A0A1B0BFC3 A0A1B0AIP9 A0A1A9X228 D3TRP1 A0A411G7P4 A0A2R5L8P8 A0A293MSY7 A0A385L4G2 A0A195E6J1 Q16QZ7 A0A195EU94 W8C765
A0A194PXV4 D9IX59 A0A3S2NM79 A0A1V1FQH9 A0A191UR67 G9C5F4 F4WPA6 A0A158NR28 E2ADN2 E9IT60 A0A2J7PX37 A0A1B6GNF1 A0A1B6D1Q1 A0A158NR27 A0A1B6I1T4 A0A3L8DWU3 R4WP03 K7J834 E2B527 A0A0T6B240 U5EZA7 E9H8M4 A0A0P4YC34 A0A1W4X1P4 A0A1Y1K6X2 A0A0M4MP46 A0A0D3RLG3 D6WYT2 A0A0P5TTR4 A0A0A9WRP1 V9IEG2 A0A087ZUP0 A0A0P6HQU0 A0A336K4X4 J3JVF7 T1DIH4 A0A1Q3EU84 T1DP10 A0A2M3YYK6 A0A2M4A1R3 W5J8H0 A0A1Y1KBX6 A0A084W0J2 A0A182IR93 A0A0K8TF46 T1II32 A0A087TZB3 A0A0P5VWS6 U4ULQ8 A0A2P8ZJQ3 A0A0K8TPL0 A0A0P6IIL4 A0A182XP04 A0A182YND2 A0A182VMX6 Q7PX08 A0A182HK46 A0A131Y0H7 B7Q5I4 A0A182Q9Y3 A0A0P5YN02 A0A224XIF0 A0A0V0G7I4 A0A023F9I8 A0A069DQV0 A0A023ENU3 A0A182MR44 A0A0P4VWN9 R4G8P2 A0A182PR85 A0A1I8NMW4 A0A0L0BZI1 A0A1L8EHB0 T1PM25 A0A1Z5L731 A0A224YU45 A0A131YYE0 L7M840 A0A131XIE5 A0A1L8DU98 A0A1A9YNT0 A0A1A9UCL9 A0A1B0BFC3 A0A1B0AIP9 A0A1A9X228 D3TRP1 A0A411G7P4 A0A2R5L8P8 A0A293MSY7 A0A385L4G2 A0A195E6J1 Q16QZ7 A0A195EU94 W8C765
PDB
3UBW
E-value=1.38099e-105,
Score=976
Ontologies
GO
PANTHER
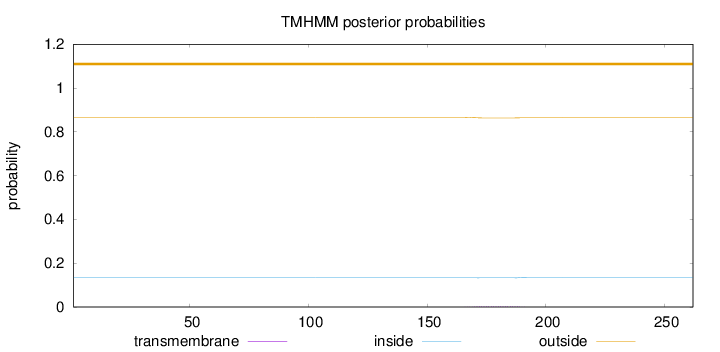
Topology
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06176
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13382
outside
1 - 262
Population Genetic Test Statistics
Pi
198.883391
Theta
189.846055
Tajima's D
0.377707
CLR
0.315621
CSRT
0.47927603619819
Interpretation
Uncertain
