Gene
KWMTBOMO09973
Pre Gene Modal
BGIBMGA013220
Annotation
PREDICTED:_estradiol_17-beta-dehydrogenase_8-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.435
Sequence
CDS
ATGAGTTTTTCTAGTAAAATCGTTTTGATTACAGGAGCCAGTAGAGGAATAGGCGCAGCAACAGCGATAATGTTCGCTAAAGAAGGGGCTGATGTAATAATTGTTGGAAGAAATGAAGAAAAACTCAAAGAAGTCGAAGTGGAATGTTCTAAATTTGGTAAGAAACCCCTTGTTATTACGGCGGATTTGGCGAAAGATGAGCAAGTTAAAGGTATAGTCAATAAAACTATAGATGCATTCGGAAAACTGGATATATTAATTAACAATGCTGGCATACTAAAGGAAAATGACATTAAGTCAGAAGGCTTCATAGAGACTTTCGACGAAGTGATGAACACAAACCTAAGAGCTTCTGTATTGATAACCAACGTGGCCATACCACACCTGATCAAAACAAGAGGCAATATCATTAATGTTTCTAGTGTCGCTGCAACTTCAACATCAGTTGTTAATTCTGTTACGTACAAAGTATCTAAAGCCGCCCTCAACCATTTCACTCGCTGCATAGCCTTAGAACTTGCTCCTCATGGCATCCGAGCTAACACAGTGAGCCCTGGCCCTGTGGAAACAGATATTTTCGAAACAGCTGGGATTCCAGAAGCACTACAAAATTATGCAGGACAACTTCCCCTTAAGAAAGTCGGCAAGAAAGAAGAAGTGGCAGATTTGATCTTGTTTTTGGCCAGCGAGAAAGCGCAAAGTATAACCGGAGCGAATTACGTTATAGATAATGGGTGGCTGTTAAACTAA
Protein
MSFSSKIVLITGASRGIGAATAIMFAKEGADVIIVGRNEEKLKEVEVECSKFGKKPLVITADLAKDEQVKGIVNKTIDAFGKLDILINNAGILKENDIKSEGFIETFDEVMNTNLRASVLITNVAIPHLIKTRGNIINVSSVAATSTSVVNSVTYKVSKAALNHFTRCIALELAPHGIRANTVSPGPVETDIFETAGIPEALQNYAGQLPLKKVGKKEEVADLILFLASEKAQSITGANYVIDNGWLLN
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JUK7
H9JUK8
A0A0N0PC39
A0A0N1IFH9
H9JUQ7
A0A2H1VC89
+ More
H9JUE7 H6AGY1 A0A2A4JIX4 H9JUQ8 A0A194PR87 A0A2A4JHN9 A0A2A4J612 A0A0N1PFW5 A0A0L7KLX4 A0A194PXT8 A0A0L7LIU8 Q9NBB5 I4DS66 A0A0N0PB24 H9JHN5 A0A194PID7 A0A0N0PAS3 A0A0L7KSS2 A0A0N0PET2 A0A194PT74 A0A2H1V3T7 A0A2H1X3T6 A0A194PJG5 A0A0N1IGV7 A0A2A4JIH1 A0A0L7KZ86 A0A0L7LH86 A0A194PIN0 A0A2A4JHX2 A0A2A4ITT1 A0A2H1VHE2 A0A2A4JIE3 A0A194PKC3 A0A2A4JH23 A0A2H1WUW2 A0A194PPC7 A0A2H1WS78 A0A2H1WTS2 A0A212EJ84 A0A212EJ88 A0A2A4J134 A0A2H1VYS0 A0A0L7LK01 A0A2A4J0E8 A0A194Q286 A0A0L7KN57 H9JUI0 A0A0N1IDX2 A0A2H1VAH5 A0A0L7KSR6 A0A2A4JIM8 D2SNW1 A0A2H1VBR9 A0A2H1WMX1 A0A2A4IZG3 A0A2A4IZU2 A0A212EP45 H9JUH9 H9JKZ2 A0A2W1BUV9 A0A0N1IHB9 A0A212F231 A0A2H1VFS6 A0A2A4K5A7 A0A0L7K599 I4DQC6 A0A0N1IIA2 A0A2H1WFW5 A0A194Q138 A0A0L7KZH2 A0A212ETR6 A0A2W1BSW8 A0A2W1BZ77 A0A2H1WPX2 A0A1Q3G5G6 A0A0L7KMQ4 A0A2H1X3K0 I4DJE1 A0A0L7KPN2 H9JG62 H9JUE8 A0A0N1PJN4 A0A1Q3G5L3 A0A1Q3G5H6 A0A194PHJ7 A0A0N0PDP5 F4WG13 A0A2A4K610 I4DPW2 A0A194RRD7 A0A1L8DJ56 A0A2H1VC15 A0A212F276 A0A194PUH3
H9JUE7 H6AGY1 A0A2A4JIX4 H9JUQ8 A0A194PR87 A0A2A4JHN9 A0A2A4J612 A0A0N1PFW5 A0A0L7KLX4 A0A194PXT8 A0A0L7LIU8 Q9NBB5 I4DS66 A0A0N0PB24 H9JHN5 A0A194PID7 A0A0N0PAS3 A0A0L7KSS2 A0A0N0PET2 A0A194PT74 A0A2H1V3T7 A0A2H1X3T6 A0A194PJG5 A0A0N1IGV7 A0A2A4JIH1 A0A0L7KZ86 A0A0L7LH86 A0A194PIN0 A0A2A4JHX2 A0A2A4ITT1 A0A2H1VHE2 A0A2A4JIE3 A0A194PKC3 A0A2A4JH23 A0A2H1WUW2 A0A194PPC7 A0A2H1WS78 A0A2H1WTS2 A0A212EJ84 A0A212EJ88 A0A2A4J134 A0A2H1VYS0 A0A0L7LK01 A0A2A4J0E8 A0A194Q286 A0A0L7KN57 H9JUI0 A0A0N1IDX2 A0A2H1VAH5 A0A0L7KSR6 A0A2A4JIM8 D2SNW1 A0A2H1VBR9 A0A2H1WMX1 A0A2A4IZG3 A0A2A4IZU2 A0A212EP45 H9JUH9 H9JKZ2 A0A2W1BUV9 A0A0N1IHB9 A0A212F231 A0A2H1VFS6 A0A2A4K5A7 A0A0L7K599 I4DQC6 A0A0N1IIA2 A0A2H1WFW5 A0A194Q138 A0A0L7KZH2 A0A212ETR6 A0A2W1BSW8 A0A2W1BZ77 A0A2H1WPX2 A0A1Q3G5G6 A0A0L7KMQ4 A0A2H1X3K0 I4DJE1 A0A0L7KPN2 H9JG62 H9JUE8 A0A0N1PJN4 A0A1Q3G5L3 A0A1Q3G5H6 A0A194PHJ7 A0A0N0PDP5 F4WG13 A0A2A4K610 I4DPW2 A0A194RRD7 A0A1L8DJ56 A0A2H1VC15 A0A212F276 A0A194PUH3
EMBL
BABH01022634
BABH01022635
BABH01022636
KQ460655
KPJ12937.1
KPJ12939.1
+ More
BABH01022835 ODYU01001522 SOQ37854.1 JF433973 AEM17060.1 NWSH01001398 PCG71373.1 KQ459595 KPI95966.1 NWSH01001353 PCG71567.1 NWSH01003146 PCG66863.1 KQ461178 KPJ07786.1 JTDY01009080 KOB64125.1 KPI95965.1 JTDY01000942 KOB75350.1 AF255341 AAF70499.1 AK405443 BAM20756.1 KPJ07785.1 BABH01012981 KQ459602 KPI93176.1 KPJ07784.1 JTDY01006084 KOB66312.1 KQ459717 KPJ20326.1 KPI95964.1 ODYU01000551 SOQ35503.1 ODYU01013212 SOQ59906.1 KPI93178.1 KPJ12938.1 PCG71374.1 JTDY01004206 KOB68460.1 JTDY01001151 KOB74729.1 KPI93177.1 PCG71376.1 NWSH01006959 PCG63175.1 ODYU01002398 SOQ39832.1 PCG71569.1 KPI93179.1 PCG71375.1 ODYU01011234 SOQ56848.1 KQ459596 KPI95296.1 ODYU01010604 SOQ55826.1 ODYU01010994 SOQ56451.1 AGBW02014512 OWR41546.1 OWR41545.1 NWSH01004416 PCG65150.1 ODYU01005275 SOQ45968.1 JTDY01000816 KOB75772.1 PCG65148.1 KQ459579 KPI99448.1 JTDY01008777 KOB64404.1 KQ460650 KPJ13113.1 SOQ37853.1 KOB66313.1 PCG71568.1 EZ407245 ACX53802.1 SOQ37852.1 ODYU01009744 SOQ54378.1 PCG65151.1 PCG65149.1 AGBW02013539 OWR43254.1 BABH01011332 KZ149910 PZC78151.1 KQ460571 KPJ13957.1 AGBW02010784 OWR47781.1 ODYU01002131 SOQ39232.1 NWSH01000134 PCG79206.1 JTDY01010313 KOB58251.1 AK404043 BAM20116.1 KQ460317 KPJ16010.1 ODYU01008068 SOQ51354.1 KQ459580 KPI99297.1 JTDY01004127 KOB68555.1 AGBW02012537 OWR44869.1 PZC78149.1 PZC78150.1 ODYU01010177 SOQ55120.1 GEYN01000194 JAV44795.1 KOB64405.1 ODYU01012735 SOQ59194.1 AK401409 BAM18031.1 JTDY01007623 KOB65070.1 BABH01002061 BABH01022833 BABH01022834 LADJ01004337 KPJ20878.1 GEYN01000185 JAV44804.1 GEYN01000184 JAV44805.1 KQ459604 KPI92523.1 KQ460254 KPJ16339.1 GL888128 EGI66780.1 NWSH01000090 PCG79695.1 AK403809 BAM19952.1 KQ459765 KPJ20037.1 GFDF01007687 JAV06397.1 ODYU01001753 SOQ38389.1 AGBW02010783 OWR47824.1 KQ459591 KPI96972.1
BABH01022835 ODYU01001522 SOQ37854.1 JF433973 AEM17060.1 NWSH01001398 PCG71373.1 KQ459595 KPI95966.1 NWSH01001353 PCG71567.1 NWSH01003146 PCG66863.1 KQ461178 KPJ07786.1 JTDY01009080 KOB64125.1 KPI95965.1 JTDY01000942 KOB75350.1 AF255341 AAF70499.1 AK405443 BAM20756.1 KPJ07785.1 BABH01012981 KQ459602 KPI93176.1 KPJ07784.1 JTDY01006084 KOB66312.1 KQ459717 KPJ20326.1 KPI95964.1 ODYU01000551 SOQ35503.1 ODYU01013212 SOQ59906.1 KPI93178.1 KPJ12938.1 PCG71374.1 JTDY01004206 KOB68460.1 JTDY01001151 KOB74729.1 KPI93177.1 PCG71376.1 NWSH01006959 PCG63175.1 ODYU01002398 SOQ39832.1 PCG71569.1 KPI93179.1 PCG71375.1 ODYU01011234 SOQ56848.1 KQ459596 KPI95296.1 ODYU01010604 SOQ55826.1 ODYU01010994 SOQ56451.1 AGBW02014512 OWR41546.1 OWR41545.1 NWSH01004416 PCG65150.1 ODYU01005275 SOQ45968.1 JTDY01000816 KOB75772.1 PCG65148.1 KQ459579 KPI99448.1 JTDY01008777 KOB64404.1 KQ460650 KPJ13113.1 SOQ37853.1 KOB66313.1 PCG71568.1 EZ407245 ACX53802.1 SOQ37852.1 ODYU01009744 SOQ54378.1 PCG65151.1 PCG65149.1 AGBW02013539 OWR43254.1 BABH01011332 KZ149910 PZC78151.1 KQ460571 KPJ13957.1 AGBW02010784 OWR47781.1 ODYU01002131 SOQ39232.1 NWSH01000134 PCG79206.1 JTDY01010313 KOB58251.1 AK404043 BAM20116.1 KQ460317 KPJ16010.1 ODYU01008068 SOQ51354.1 KQ459580 KPI99297.1 JTDY01004127 KOB68555.1 AGBW02012537 OWR44869.1 PZC78149.1 PZC78150.1 ODYU01010177 SOQ55120.1 GEYN01000194 JAV44795.1 KOB64405.1 ODYU01012735 SOQ59194.1 AK401409 BAM18031.1 JTDY01007623 KOB65070.1 BABH01002061 BABH01022833 BABH01022834 LADJ01004337 KPJ20878.1 GEYN01000185 JAV44804.1 GEYN01000184 JAV44805.1 KQ459604 KPI92523.1 KQ460254 KPJ16339.1 GL888128 EGI66780.1 NWSH01000090 PCG79695.1 AK403809 BAM19952.1 KQ459765 KPJ20037.1 GFDF01007687 JAV06397.1 ODYU01001753 SOQ38389.1 AGBW02010783 OWR47824.1 KQ459591 KPI96972.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JUK7
H9JUK8
A0A0N0PC39
A0A0N1IFH9
H9JUQ7
A0A2H1VC89
+ More
H9JUE7 H6AGY1 A0A2A4JIX4 H9JUQ8 A0A194PR87 A0A2A4JHN9 A0A2A4J612 A0A0N1PFW5 A0A0L7KLX4 A0A194PXT8 A0A0L7LIU8 Q9NBB5 I4DS66 A0A0N0PB24 H9JHN5 A0A194PID7 A0A0N0PAS3 A0A0L7KSS2 A0A0N0PET2 A0A194PT74 A0A2H1V3T7 A0A2H1X3T6 A0A194PJG5 A0A0N1IGV7 A0A2A4JIH1 A0A0L7KZ86 A0A0L7LH86 A0A194PIN0 A0A2A4JHX2 A0A2A4ITT1 A0A2H1VHE2 A0A2A4JIE3 A0A194PKC3 A0A2A4JH23 A0A2H1WUW2 A0A194PPC7 A0A2H1WS78 A0A2H1WTS2 A0A212EJ84 A0A212EJ88 A0A2A4J134 A0A2H1VYS0 A0A0L7LK01 A0A2A4J0E8 A0A194Q286 A0A0L7KN57 H9JUI0 A0A0N1IDX2 A0A2H1VAH5 A0A0L7KSR6 A0A2A4JIM8 D2SNW1 A0A2H1VBR9 A0A2H1WMX1 A0A2A4IZG3 A0A2A4IZU2 A0A212EP45 H9JUH9 H9JKZ2 A0A2W1BUV9 A0A0N1IHB9 A0A212F231 A0A2H1VFS6 A0A2A4K5A7 A0A0L7K599 I4DQC6 A0A0N1IIA2 A0A2H1WFW5 A0A194Q138 A0A0L7KZH2 A0A212ETR6 A0A2W1BSW8 A0A2W1BZ77 A0A2H1WPX2 A0A1Q3G5G6 A0A0L7KMQ4 A0A2H1X3K0 I4DJE1 A0A0L7KPN2 H9JG62 H9JUE8 A0A0N1PJN4 A0A1Q3G5L3 A0A1Q3G5H6 A0A194PHJ7 A0A0N0PDP5 F4WG13 A0A2A4K610 I4DPW2 A0A194RRD7 A0A1L8DJ56 A0A2H1VC15 A0A212F276 A0A194PUH3
H9JUE7 H6AGY1 A0A2A4JIX4 H9JUQ8 A0A194PR87 A0A2A4JHN9 A0A2A4J612 A0A0N1PFW5 A0A0L7KLX4 A0A194PXT8 A0A0L7LIU8 Q9NBB5 I4DS66 A0A0N0PB24 H9JHN5 A0A194PID7 A0A0N0PAS3 A0A0L7KSS2 A0A0N0PET2 A0A194PT74 A0A2H1V3T7 A0A2H1X3T6 A0A194PJG5 A0A0N1IGV7 A0A2A4JIH1 A0A0L7KZ86 A0A0L7LH86 A0A194PIN0 A0A2A4JHX2 A0A2A4ITT1 A0A2H1VHE2 A0A2A4JIE3 A0A194PKC3 A0A2A4JH23 A0A2H1WUW2 A0A194PPC7 A0A2H1WS78 A0A2H1WTS2 A0A212EJ84 A0A212EJ88 A0A2A4J134 A0A2H1VYS0 A0A0L7LK01 A0A2A4J0E8 A0A194Q286 A0A0L7KN57 H9JUI0 A0A0N1IDX2 A0A2H1VAH5 A0A0L7KSR6 A0A2A4JIM8 D2SNW1 A0A2H1VBR9 A0A2H1WMX1 A0A2A4IZG3 A0A2A4IZU2 A0A212EP45 H9JUH9 H9JKZ2 A0A2W1BUV9 A0A0N1IHB9 A0A212F231 A0A2H1VFS6 A0A2A4K5A7 A0A0L7K599 I4DQC6 A0A0N1IIA2 A0A2H1WFW5 A0A194Q138 A0A0L7KZH2 A0A212ETR6 A0A2W1BSW8 A0A2W1BZ77 A0A2H1WPX2 A0A1Q3G5G6 A0A0L7KMQ4 A0A2H1X3K0 I4DJE1 A0A0L7KPN2 H9JG62 H9JUE8 A0A0N1PJN4 A0A1Q3G5L3 A0A1Q3G5H6 A0A194PHJ7 A0A0N0PDP5 F4WG13 A0A2A4K610 I4DPW2 A0A194RRD7 A0A1L8DJ56 A0A2H1VC15 A0A212F276 A0A194PUH3
PDB
1XHL
E-value=7.02364e-37,
Score=383
Ontologies
Topology
Subcellular location
Nucleus
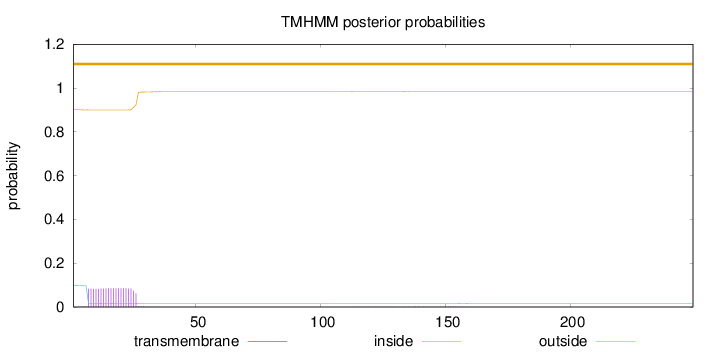
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.71076
Exp number, first 60 AAs:
1.67843
Total prob of N-in:
0.09952
outside
1 - 249
Population Genetic Test Statistics
Pi
210.090473
Theta
185.036447
Tajima's D
0.440662
CLR
0.071351
CSRT
0.494475276236188
Interpretation
Uncertain
