Pre Gene Modal
BGIBMGA013223
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup88_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.945
Sequence
CDS
ATGAGCAGCTGTGGTAGCAGAATCTGCGTCTGGGGTACTCAAGGTGTCACTATAGCTGAACTACCTTCGAGATGGGGTCGCGGTGGACTCTTCGATTCTGGCAGTCAAACCGTGCTTTGCAAGAGCTATAGTCTGGACGAGAAATTCCTGTACACGACTGGCGAGATTCGCAGGGTCCACTGGCATCCGATATCTCTATCACACGTACTGGTCTTAGTCTCCAATAATGCGATCAGACTGTACAATGTAACCTTGAAAACCGGTCCGAAGCTGGTGAAGACATACTCTATTGGTCCGAAACCGACCAGCCTCCTCGCCGGCAAGACGATTCTGGACAGTCTCGGAGACACTGCAGTGGATTTCACGCCGACGCCTGACGCGGAGCACATCCTGATCCTGAGGGGCGATGGAGAAATTTACATGATGGACTGTGATCTGACTAATAAAAGTCCACTCCAGCCGAAGCTCGTTGGACCCCTCGCCATATATCCCCCTGCGGACGACAATTACGGCTCGGACTCTTGCTGCATCCTGTGCATGGGGGGCAGCGACATCCCCCCCCTGGTCGTCATCGCGACTTCCTCGGCCGCTCTGTACCACTGCTTGCTGCTACCAAATAGCGAGAAAGAAGAGAGCGACAGAGACGGCTACGCTCTGTACGTGGTGGAGACCGTGGAGTTGGACGTGGTACCGGAGCCCGACGCGGAGCCCTACCCCGTGCAGCTGATCAAGTGCACGGACGACACGTACGCGTGCGTGCACGCGGCGGGCGCGCACACCGTCGCGCTGCCCGTGCTCGCCGCGCTGCGACACTACGCGCGCGCACCCGACGGTAACCACCCGCCGCTCGGTCGGCTGTACGGACACACCACACTCACTGTCCACTCGCCGCTCTGTCGGCTGTAG
Protein
MSSCGSRICVWGTQGVTIAELPSRWGRGGLFDSGSQTVLCKSYSLDEKFLYTTGEIRRVHWHPISLSHVLVLVSNNAIRLYNVTLKTGPKLVKTYSIGPKPTSLLAGKTILDSLGDTAVDFTPTPDAEHILILRGDGEIYMMDCDLTNKSPLQPKLVGPLAIYPPADDNYGSDSCCILCMGGSDIPPLVVIATSSAALYHCLLLPNSEKEESDRDGYALYVVETVELDVVPEPDAEPYPVQLIKCTDDTYACVHAAGAHTVALPVLAALRHYARAPDGNHPPLGRLYGHTTLTVHSPLCRL
Summary
Uniprot
H9JUL0
A0A2H1V4V0
A0A2A4J2J7
A0A437B4U6
A0A194PR82
A0A212F238
+ More
A0A088AVE5 A0A2A3EA02 C3Z4B2 A0A3L8DCT3 A0A067RBL6 A0A195C450 A0A0L7R3E1 A0A026WS47 A0A158NAP1 A0A195FSH0 K7IRP9 A0A2G8L9L5 A0A195ELF7 A0A195BK04 F4WC99 A0A2C9LBJ3 A0A232FAS0 A0A2H8THQ2 A0A2S2QPX8 J9JUX4 A0A1S3K2A1 A0A091RBY0 A0A210PUU6 A0A091FZK2 A0A091KVE7 A0A226NAZ1 F7C806 A0A226PFM8 A0A2J7PF88 A0A091M9F2 R0KBU3 U3J887 A0A0M9A1Q9 A0A2J7PF85 A0A2J7PF78 Q4KLQ6 A0A1L8HGG3 A0A091VFQ6 Q707N0 Q6DDV7 A0A091LLF1 A0A099ZN49 A0A1W7RF41 A0A3M6TZT3 A0A0F7Z6N7 A0A2I0M1N3 A0A1V4K4F1 A0A2P4T6F4 A0A091QYZ7 A0A091F5W4 A0A401RK05 A0A093HMK8 A0A091H560 A0A0A0AIR4 G1MW57 A0A093NUJ0 A0A087QWL4 A0A091JKJ7 A0A2B4SJ52 A0A3S3QC56 A0A091WBI7 A0A287BTG5 A0A091TCL3 A0A091SQC6 A0A2L2Y686 A0A093TM60 A0A093FQV0 A0A1L8H8E3 Q707M9 A0A287A9U8 A0A093EX49 A0A093QBM5 A0A094KUV1 A0A3B4CFQ5 A0A310SHE4 V8NR27 A0A093LQS9 F6Y295 G9KEE4 A0A2R8QUY6 H0Z1I3 A0A3B1JBP6 A0A1S3EXT6 A0A093G5N5 M3X5U7 A0A3B1JX91 E2BG54 A2CEI4 W5KJA4 A0A087TNT3 A0A444V329
A0A088AVE5 A0A2A3EA02 C3Z4B2 A0A3L8DCT3 A0A067RBL6 A0A195C450 A0A0L7R3E1 A0A026WS47 A0A158NAP1 A0A195FSH0 K7IRP9 A0A2G8L9L5 A0A195ELF7 A0A195BK04 F4WC99 A0A2C9LBJ3 A0A232FAS0 A0A2H8THQ2 A0A2S2QPX8 J9JUX4 A0A1S3K2A1 A0A091RBY0 A0A210PUU6 A0A091FZK2 A0A091KVE7 A0A226NAZ1 F7C806 A0A226PFM8 A0A2J7PF88 A0A091M9F2 R0KBU3 U3J887 A0A0M9A1Q9 A0A2J7PF85 A0A2J7PF78 Q4KLQ6 A0A1L8HGG3 A0A091VFQ6 Q707N0 Q6DDV7 A0A091LLF1 A0A099ZN49 A0A1W7RF41 A0A3M6TZT3 A0A0F7Z6N7 A0A2I0M1N3 A0A1V4K4F1 A0A2P4T6F4 A0A091QYZ7 A0A091F5W4 A0A401RK05 A0A093HMK8 A0A091H560 A0A0A0AIR4 G1MW57 A0A093NUJ0 A0A087QWL4 A0A091JKJ7 A0A2B4SJ52 A0A3S3QC56 A0A091WBI7 A0A287BTG5 A0A091TCL3 A0A091SQC6 A0A2L2Y686 A0A093TM60 A0A093FQV0 A0A1L8H8E3 Q707M9 A0A287A9U8 A0A093EX49 A0A093QBM5 A0A094KUV1 A0A3B4CFQ5 A0A310SHE4 V8NR27 A0A093LQS9 F6Y295 G9KEE4 A0A2R8QUY6 H0Z1I3 A0A3B1JBP6 A0A1S3EXT6 A0A093G5N5 M3X5U7 A0A3B1JX91 E2BG54 A2CEI4 W5KJA4 A0A087TNT3 A0A444V329
Pubmed
EMBL
BABH01022652
ODYU01000679
SOQ35863.1
NWSH01003392
PCG66377.1
RSAL01000158
+ More
RVE45530.1 KQ459595 KPI95961.1 AGBW02010784 OWR47784.1 KZ288311 PBC28555.1 GG666578 EEN52725.1 QOIP01000009 RLU18310.1 KK852571 KDR21117.1 KQ978292 KYM95395.1 KQ414663 KOC65343.1 KK107119 EZA58852.1 ADTU01009895 ADTU01009896 ADTU01009897 ADTU01009898 ADTU01009899 KQ981305 KYN42849.1 MRZV01000159 PIK56946.1 KQ978739 KYN28767.1 KQ976455 KYM85027.1 GL888070 EGI68079.1 NNAY01000508 OXU27941.1 GFXV01001848 MBW13653.1 GGMS01010377 MBY79580.1 ABLF02022558 KK812755 KFQ36757.1 NEDP02005477 OWF40258.1 KL447643 KFO75645.1 KK543951 KFP31719.1 MCFN01000112 OXB64754.1 AAMC01063017 AWGT02000078 OXB78866.1 NEVH01026085 PNF14995.1 KK526789 KFP68438.1 KB742489 EOB07826.1 ADON01011640 KQ435762 KOX75540.1 PNF14994.1 PNF14993.1 BC099049 AAH99049.1 CM004468 OCT95176.1 KL410945 KFR01288.1 AJ617672 CAE85030.1 BC077397 AAH77397.1 KK756889 KFP43631.1 KL896091 KGL83207.1 GDAY02001663 JAV49764.1 RCHS01002548 RMX46867.1 GBEX01002324 JAI12236.1 AKCR02000048 PKK23585.1 LSYS01005108 OPJ78757.1 PPHD01007173 POI31943.1 KK685972 KFQ14574.1 KK719555 KFO64247.1 BEZZ01002969 GCC18488.1 KL206523 KFV82971.1 KL524072 KFO90549.1 KL871755 KGL93348.1 KL224795 KFW63812.1 KL225959 KFM05618.1 KK502172 KFP21117.1 LSMT01000082 PFX28502.1 NCKU01000064 RWS17522.1 KK733763 KFQ99090.1 AEMK02000082 KK447438 KFQ72059.1 KK482465 KFQ60917.1 IAAA01018538 LAA03674.1 KL445521 KFW95339.1 KK632805 KFV56686.1 CM004469 OCT92377.1 AJ617673 CAE85031.1 KK377050 KFV46469.1 KL672574 KFW86373.1 KL267256 KFZ62405.1 KQ763450 OAD55069.1 AZIM01002217 ETE64505.1 KK576996 KFW11071.1 JP014671 AES03269.1 BX957294 ABQF01026138 KL215363 KFV64398.1 AANG04000740 GL448115 EFN85322.1 CR450697 KK116094 KFM66772.1 SCEB01002963 RXM94817.1
RVE45530.1 KQ459595 KPI95961.1 AGBW02010784 OWR47784.1 KZ288311 PBC28555.1 GG666578 EEN52725.1 QOIP01000009 RLU18310.1 KK852571 KDR21117.1 KQ978292 KYM95395.1 KQ414663 KOC65343.1 KK107119 EZA58852.1 ADTU01009895 ADTU01009896 ADTU01009897 ADTU01009898 ADTU01009899 KQ981305 KYN42849.1 MRZV01000159 PIK56946.1 KQ978739 KYN28767.1 KQ976455 KYM85027.1 GL888070 EGI68079.1 NNAY01000508 OXU27941.1 GFXV01001848 MBW13653.1 GGMS01010377 MBY79580.1 ABLF02022558 KK812755 KFQ36757.1 NEDP02005477 OWF40258.1 KL447643 KFO75645.1 KK543951 KFP31719.1 MCFN01000112 OXB64754.1 AAMC01063017 AWGT02000078 OXB78866.1 NEVH01026085 PNF14995.1 KK526789 KFP68438.1 KB742489 EOB07826.1 ADON01011640 KQ435762 KOX75540.1 PNF14994.1 PNF14993.1 BC099049 AAH99049.1 CM004468 OCT95176.1 KL410945 KFR01288.1 AJ617672 CAE85030.1 BC077397 AAH77397.1 KK756889 KFP43631.1 KL896091 KGL83207.1 GDAY02001663 JAV49764.1 RCHS01002548 RMX46867.1 GBEX01002324 JAI12236.1 AKCR02000048 PKK23585.1 LSYS01005108 OPJ78757.1 PPHD01007173 POI31943.1 KK685972 KFQ14574.1 KK719555 KFO64247.1 BEZZ01002969 GCC18488.1 KL206523 KFV82971.1 KL524072 KFO90549.1 KL871755 KGL93348.1 KL224795 KFW63812.1 KL225959 KFM05618.1 KK502172 KFP21117.1 LSMT01000082 PFX28502.1 NCKU01000064 RWS17522.1 KK733763 KFQ99090.1 AEMK02000082 KK447438 KFQ72059.1 KK482465 KFQ60917.1 IAAA01018538 LAA03674.1 KL445521 KFW95339.1 KK632805 KFV56686.1 CM004469 OCT92377.1 AJ617673 CAE85031.1 KK377050 KFV46469.1 KL672574 KFW86373.1 KL267256 KFZ62405.1 KQ763450 OAD55069.1 AZIM01002217 ETE64505.1 KK576996 KFW11071.1 JP014671 AES03269.1 BX957294 ABQF01026138 KL215363 KFV64398.1 AANG04000740 GL448115 EFN85322.1 CR450697 KK116094 KFM66772.1 SCEB01002963 RXM94817.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000005203
+ More
UP000242457 UP000001554 UP000279307 UP000027135 UP000078542 UP000053825 UP000053097 UP000005205 UP000078541 UP000002358 UP000230750 UP000078492 UP000078540 UP000007755 UP000076420 UP000215335 UP000007819 UP000085678 UP000242188 UP000053760 UP000198323 UP000008143 UP000198419 UP000235965 UP000016666 UP000053105 UP000186698 UP000053283 UP000053641 UP000275408 UP000053872 UP000190648 UP000052976 UP000287033 UP000053584 UP000053858 UP000001645 UP000054081 UP000053286 UP000053119 UP000225706 UP000285301 UP000053605 UP000008227 UP000053258 UP000261440 UP000000437 UP000007754 UP000018467 UP000081671 UP000053875 UP000011712 UP000008237 UP000054359
UP000242457 UP000001554 UP000279307 UP000027135 UP000078542 UP000053825 UP000053097 UP000005205 UP000078541 UP000002358 UP000230750 UP000078492 UP000078540 UP000007755 UP000076420 UP000215335 UP000007819 UP000085678 UP000242188 UP000053760 UP000198323 UP000008143 UP000198419 UP000235965 UP000016666 UP000053105 UP000186698 UP000053283 UP000053641 UP000275408 UP000053872 UP000190648 UP000052976 UP000287033 UP000053584 UP000053858 UP000001645 UP000054081 UP000053286 UP000053119 UP000225706 UP000285301 UP000053605 UP000008227 UP000053258 UP000261440 UP000000437 UP000007754 UP000018467 UP000081671 UP000053875 UP000011712 UP000008237 UP000054359
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JUL0
A0A2H1V4V0
A0A2A4J2J7
A0A437B4U6
A0A194PR82
A0A212F238
+ More
A0A088AVE5 A0A2A3EA02 C3Z4B2 A0A3L8DCT3 A0A067RBL6 A0A195C450 A0A0L7R3E1 A0A026WS47 A0A158NAP1 A0A195FSH0 K7IRP9 A0A2G8L9L5 A0A195ELF7 A0A195BK04 F4WC99 A0A2C9LBJ3 A0A232FAS0 A0A2H8THQ2 A0A2S2QPX8 J9JUX4 A0A1S3K2A1 A0A091RBY0 A0A210PUU6 A0A091FZK2 A0A091KVE7 A0A226NAZ1 F7C806 A0A226PFM8 A0A2J7PF88 A0A091M9F2 R0KBU3 U3J887 A0A0M9A1Q9 A0A2J7PF85 A0A2J7PF78 Q4KLQ6 A0A1L8HGG3 A0A091VFQ6 Q707N0 Q6DDV7 A0A091LLF1 A0A099ZN49 A0A1W7RF41 A0A3M6TZT3 A0A0F7Z6N7 A0A2I0M1N3 A0A1V4K4F1 A0A2P4T6F4 A0A091QYZ7 A0A091F5W4 A0A401RK05 A0A093HMK8 A0A091H560 A0A0A0AIR4 G1MW57 A0A093NUJ0 A0A087QWL4 A0A091JKJ7 A0A2B4SJ52 A0A3S3QC56 A0A091WBI7 A0A287BTG5 A0A091TCL3 A0A091SQC6 A0A2L2Y686 A0A093TM60 A0A093FQV0 A0A1L8H8E3 Q707M9 A0A287A9U8 A0A093EX49 A0A093QBM5 A0A094KUV1 A0A3B4CFQ5 A0A310SHE4 V8NR27 A0A093LQS9 F6Y295 G9KEE4 A0A2R8QUY6 H0Z1I3 A0A3B1JBP6 A0A1S3EXT6 A0A093G5N5 M3X5U7 A0A3B1JX91 E2BG54 A2CEI4 W5KJA4 A0A087TNT3 A0A444V329
A0A088AVE5 A0A2A3EA02 C3Z4B2 A0A3L8DCT3 A0A067RBL6 A0A195C450 A0A0L7R3E1 A0A026WS47 A0A158NAP1 A0A195FSH0 K7IRP9 A0A2G8L9L5 A0A195ELF7 A0A195BK04 F4WC99 A0A2C9LBJ3 A0A232FAS0 A0A2H8THQ2 A0A2S2QPX8 J9JUX4 A0A1S3K2A1 A0A091RBY0 A0A210PUU6 A0A091FZK2 A0A091KVE7 A0A226NAZ1 F7C806 A0A226PFM8 A0A2J7PF88 A0A091M9F2 R0KBU3 U3J887 A0A0M9A1Q9 A0A2J7PF85 A0A2J7PF78 Q4KLQ6 A0A1L8HGG3 A0A091VFQ6 Q707N0 Q6DDV7 A0A091LLF1 A0A099ZN49 A0A1W7RF41 A0A3M6TZT3 A0A0F7Z6N7 A0A2I0M1N3 A0A1V4K4F1 A0A2P4T6F4 A0A091QYZ7 A0A091F5W4 A0A401RK05 A0A093HMK8 A0A091H560 A0A0A0AIR4 G1MW57 A0A093NUJ0 A0A087QWL4 A0A091JKJ7 A0A2B4SJ52 A0A3S3QC56 A0A091WBI7 A0A287BTG5 A0A091TCL3 A0A091SQC6 A0A2L2Y686 A0A093TM60 A0A093FQV0 A0A1L8H8E3 Q707M9 A0A287A9U8 A0A093EX49 A0A093QBM5 A0A094KUV1 A0A3B4CFQ5 A0A310SHE4 V8NR27 A0A093LQS9 F6Y295 G9KEE4 A0A2R8QUY6 H0Z1I3 A0A3B1JBP6 A0A1S3EXT6 A0A093G5N5 M3X5U7 A0A3B1JX91 E2BG54 A2CEI4 W5KJA4 A0A087TNT3 A0A444V329
Ontologies
KEGG
GO
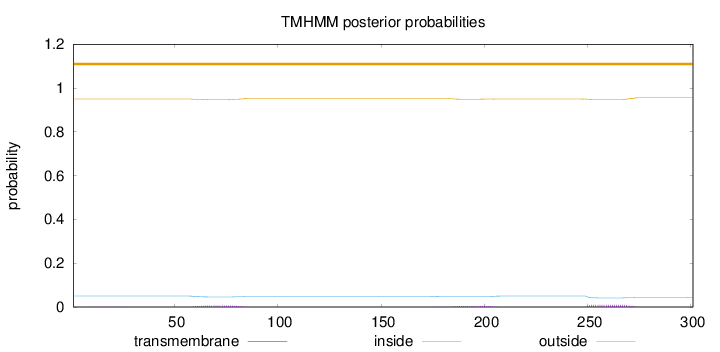
Topology
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44032
Exp number, first 60 AAs:
0.01128
Total prob of N-in:
0.05032
outside
1 - 301
Population Genetic Test Statistics
Pi
239.693574
Theta
171.422492
Tajima's D
1.181127
CLR
0.365816
CSRT
0.715814209289536
Interpretation
Uncertain
