Pre Gene Modal
BGIBMGA013190
Annotation
PREDICTED:_vacuolar_protein_sorting_29_isoform_X1_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 29
Alternative Name
Vesicle protein sorting 29
Location in the cell
Cytoplasmic Reliability : 1.841
Sequence
CDS
ATGTTGGTACTTGTCCTCGGGGATCTTCACATTCCTCACAGATGCAGCAGCTTGCCAGCTAAGTTCAAGAAGTTGCTTCTACCTGGAAGGATACAGCATATATTGTGTACTGGAAACTTATGTACAAAAGACTCCTATGATTATTTGAAGACTTTGGCCAGTGATGTGCATGTTGTTAGGGGAGATTTTGATGAGAATGCGACATATCCAGAGCAGAAAGTCGTCACAGTTGGACAGTTCCGCATTGGACTGATTCATGGACACCAAGTAGTCCCTTGGGGCGATGAAGAGTCTCTAGCTTTGATACAGAGGCAGCTGGATGTGGACATCCTGATATCAGGGCACACGCATCGCTTTGAGGCTTACGAACACGAGAATAAGTTCTATATCAATCCTGGTTCAGCTACTGGAGGTTACAGCCCTTTATACAGGGATCCTACTCCTTCGTTTGTGCTGATGGACATTCAGAGTTCGACAGTGGTCACCTACGTTTACAAGCTCCTCGGTGATGAAGTCAAAGTTGAAAGAATTGAATACAAGAAGACATGA
Protein
MLVLVLGDLHIPHRCSSLPAKFKKLLLPGRIQHILCTGNLCTKDSYDYLKTLASDVHVVRGDFDENATYPEQKVVTVGQFRIGLIHGHQVVPWGDEESLALIQRQLDVDILISGHTHRFEAYEHENKFYINPGSATGGYSPLYRDPTPSFVLMDIQSSTVVTYVYKLLGDEVKVERIEYKKT
Summary
Description
Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway.
Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. Retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) (By similarity). Acts also as component of the retriever complex. The retriever complex is an heterotrimeric complex related to retromer cargo-selective complex (CSC) and essential for retromer-independent retrieval and recycling of numerous cargos such as integrins. In the endosomes, retriever complex drives the retrieval and recycling of NxxY-motif-containing cargo proteins by coupling to SNX17, a cargo essential for the homeostatic maintenance of numerous dell suface proteins associated with processes that include cell migration, cell adhesion, nutrient supply and cell signaling (By similarity).
Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. Retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) (By similarity). Acts also as component of the retriever complex. The retriever complex is an heterotrimeric complex related to retromer cargo-selective complex (CSC) and essential for retromer-independent retrieval and recycling of numerous cargos such as integrins. In the endosomes, retriever complex drives the retrieval and recycling of NxxY-motif-containing cargo proteins by coupling to SNX17, a cargo essential for the homeostatic maintenance of numerous dell suface proteins associated with processes that include cell migration, cell adhesion, nutrient supply and cell signaling (By similarity).
Subunit
Component of the heterotrimeric retromer cargo-selective complex (CSC) which is believed to associate with variable sorting nexins to form functionally distinct retromer complex variants (By similarity). Component of the heterotrimeric retriever complex (By similarity).
Similarity
Belongs to the VPS29 family.
Keywords
Complete proteome
Cytoplasm
Endosome
Membrane
Metal-binding
Protein transport
Reference proteome
Transport
Zinc
Feature
chain Vacuolar protein sorting-associated protein 29
Uniprot
H9JUH7
Q1HPG7
I4DQC7
A0A3S2P9Y1
A0A2A4J009
S4PJX1
+ More
A0A212F235 A0A0N1PIZ8 A0A195BVK4 A0A151WSX7 A0A026VWC9 A0A0L7QPQ8 A0A2A3E7G3 A0A195FQC0 A0A0M8ZUJ4 A0A1W4XM03 A0A1S4N1Z2 A0A158NV20 F4WXI8 A0A1B6KBG3 A0A1B6IRK5 A0A1B6EIF4 R4V4Z7 A0A1B6EA34 A0A224YT62 A0A131YGQ7 L7M701 A0A131XBB0 K7ITM8 A0A1A7WMD7 A0A067RKM6 A0A1A8D8K1 A0A1A8KNE5 A0A1A8UC64 A0A1A8SDI7 A0A1A8Q0G0 A0A1Y1KZP6 A0A2R5LLT0 A0A1A7YQ83 A0A023GIR9 G3MMP1 A0A023FIU7 A0A023FUV0 A0A1E1WZN2 A0A1Z5KUA4 D2A6E5 T1DGE8 A0A2M3Z8R8 A0A2M4ANY8 W5JEV3 A0A182FR85 A0A293LA52 A0A087URV3 J3JXW3 B7P164 A0A0K8RNP5 A0A182LTS8 A0A182RNL6 A0A182N4L6 A0A182PZU0 A0A182Y7B1 A0A182K5M4 A0A182VQ81 Q7ZV68 A0A1A8CWC6 A0A1A8AV03 A0A1A8R307 A0A182PTH3 A0A182VHK4 A0A182TUD7 A0A182XEI3 A0A182LFI1 Q7QBW4 A0A182HTC9 A0A2I4CC29 A0A402F869 A0A3Q3B3M9 A0A2M4AQ85 A0A3Q3KSW7 A0A182IZA4 A0A3Q2P9J9 A0A3B3Y244 A0A0S7LYW6 A0A3B5RA41 A0A3B3D2V0 A0A3B3UAZ5 A0A3P9NFP8 H2MUI2 A0A3S2PPD7 A0A3P9KV97 A0A2R7WIG8 A0A084VE18 A0A0P4VUV8 R4G562 A0A195CCK9 A0A3L8SD62 A0A218V657 A0A3M0KF96 A0A226PZG7 A0A1V4JS14
A0A212F235 A0A0N1PIZ8 A0A195BVK4 A0A151WSX7 A0A026VWC9 A0A0L7QPQ8 A0A2A3E7G3 A0A195FQC0 A0A0M8ZUJ4 A0A1W4XM03 A0A1S4N1Z2 A0A158NV20 F4WXI8 A0A1B6KBG3 A0A1B6IRK5 A0A1B6EIF4 R4V4Z7 A0A1B6EA34 A0A224YT62 A0A131YGQ7 L7M701 A0A131XBB0 K7ITM8 A0A1A7WMD7 A0A067RKM6 A0A1A8D8K1 A0A1A8KNE5 A0A1A8UC64 A0A1A8SDI7 A0A1A8Q0G0 A0A1Y1KZP6 A0A2R5LLT0 A0A1A7YQ83 A0A023GIR9 G3MMP1 A0A023FIU7 A0A023FUV0 A0A1E1WZN2 A0A1Z5KUA4 D2A6E5 T1DGE8 A0A2M3Z8R8 A0A2M4ANY8 W5JEV3 A0A182FR85 A0A293LA52 A0A087URV3 J3JXW3 B7P164 A0A0K8RNP5 A0A182LTS8 A0A182RNL6 A0A182N4L6 A0A182PZU0 A0A182Y7B1 A0A182K5M4 A0A182VQ81 Q7ZV68 A0A1A8CWC6 A0A1A8AV03 A0A1A8R307 A0A182PTH3 A0A182VHK4 A0A182TUD7 A0A182XEI3 A0A182LFI1 Q7QBW4 A0A182HTC9 A0A2I4CC29 A0A402F869 A0A3Q3B3M9 A0A2M4AQ85 A0A3Q3KSW7 A0A182IZA4 A0A3Q2P9J9 A0A3B3Y244 A0A0S7LYW6 A0A3B5RA41 A0A3B3D2V0 A0A3B3UAZ5 A0A3P9NFP8 H2MUI2 A0A3S2PPD7 A0A3P9KV97 A0A2R7WIG8 A0A084VE18 A0A0P4VUV8 R4G562 A0A195CCK9 A0A3L8SD62 A0A218V657 A0A3M0KF96 A0A226PZG7 A0A1V4JS14
Pubmed
19121390
22651552
23622113
22118469
26354079
24508170
+ More
30249741 20566863 21347285 21719571 28797301 26830274 25576852 28049606 20075255 24845553 28004739 22216098 28503490 28528879 18362917 19820115 20920257 23761445 22516182 25244985 23594743 20966253 12364791 14747013 17210077 23542700 29451363 17554307 24438588 27129103 30282656 24621616
30249741 20566863 21347285 21719571 28797301 26830274 25576852 28049606 20075255 24845553 28004739 22216098 28503490 28528879 18362917 19820115 20920257 23761445 22516182 25244985 23594743 20966253 12364791 14747013 17210077 23542700 29451363 17554307 24438588 27129103 30282656 24621616
EMBL
BABH01022658
DQ443435
ABF51524.1
AK404044
BAM20117.1
RSAL01000158
+ More
RVE45532.1 NWSH01004688 PCG64770.1 GAIX01004780 JAA87780.1 AGBW02010784 OWR47786.1 KQ460655 KPJ12943.1 KQ976401 KYM92612.1 KQ982769 KYQ50897.1 KK107727 QOIP01000010 EZA47975.1 RLU17914.1 KQ414813 KOC60544.1 KZ288344 PBC27687.1 KQ981335 KYN42616.1 KQ435840 KOX71425.1 AAZO01007430 ADTU01026857 GL888425 EGI61058.1 GEBQ01031180 JAT08797.1 GECU01018176 JAS89530.1 GECZ01032071 GECZ01009762 JAS37698.1 JAS60007.1 KC741109 AGM32933.1 GEDC01002507 JAS34791.1 GFPF01009641 MAA20787.1 GEDV01010852 JAP77705.1 GACK01006096 JAA58938.1 GEFH01004372 JAP64209.1 HADW01005366 SBP06766.1 KK852571 KDR21120.1 HAEA01001972 SBQ30452.1 HAEE01013900 SBR33950.1 HAEJ01004256 SBS44713.1 HAEI01013141 SBS15610.1 HAEF01011412 HAEG01010485 SBR87250.1 GEZM01068657 GEZM01068656 GEZM01068655 JAV66824.1 GGLE01006323 MBY10449.1 HADX01010106 SBP32338.1 GBBM01002753 JAC32665.1 JO843142 AEO34759.1 GBBK01003929 JAC20553.1 GBBL01002061 JAC25259.1 GFAC01006942 JAT92246.1 GFJQ02008288 JAV98681.1 KQ971347 EFA05502.1 GAMD01002801 JAA98789.1 GGFM01004129 MBW24880.1 GGFK01009131 MBW42452.1 ADMH02001603 GGFL01004223 ETN61873.1 MBW68401.1 GFWV01005447 MAA30177.1 KK121265 KFM80092.1 BT128085 AEE63046.1 ABJB010872459 DS615069 EEC00336.1 GADI01001594 GEFM01003767 JAA72214.1 JAP72029.1 AXCM01003145 AXCN02000047 AL844518 BC045981 BC071331 HADZ01020103 SBP84044.1 HADY01020028 SBP58513.1 HAEH01014538 SBS00351.1 AAAB01008859 EAA08218.2 APCN01000459 BDOT01000138 GCF52634.1 GGFK01009608 MBW42929.1 GBYX01137196 JAO94622.1 CM012447 RVE66327.1 KK854881 PTY19444.1 ATLV01012143 KE524764 KFB36212.1 GDKW01000775 JAI55820.1 ACPB03015836 GAHY01000775 JAA76735.1 KQ978023 KYM97961.1 QUSF01000030 RLV99829.1 MUZQ01000045 OWK61161.1 QRBI01000121 RMC05787.1 AWGT02000001 OXB84910.1 LSYS01006629 OPJ75016.1
RVE45532.1 NWSH01004688 PCG64770.1 GAIX01004780 JAA87780.1 AGBW02010784 OWR47786.1 KQ460655 KPJ12943.1 KQ976401 KYM92612.1 KQ982769 KYQ50897.1 KK107727 QOIP01000010 EZA47975.1 RLU17914.1 KQ414813 KOC60544.1 KZ288344 PBC27687.1 KQ981335 KYN42616.1 KQ435840 KOX71425.1 AAZO01007430 ADTU01026857 GL888425 EGI61058.1 GEBQ01031180 JAT08797.1 GECU01018176 JAS89530.1 GECZ01032071 GECZ01009762 JAS37698.1 JAS60007.1 KC741109 AGM32933.1 GEDC01002507 JAS34791.1 GFPF01009641 MAA20787.1 GEDV01010852 JAP77705.1 GACK01006096 JAA58938.1 GEFH01004372 JAP64209.1 HADW01005366 SBP06766.1 KK852571 KDR21120.1 HAEA01001972 SBQ30452.1 HAEE01013900 SBR33950.1 HAEJ01004256 SBS44713.1 HAEI01013141 SBS15610.1 HAEF01011412 HAEG01010485 SBR87250.1 GEZM01068657 GEZM01068656 GEZM01068655 JAV66824.1 GGLE01006323 MBY10449.1 HADX01010106 SBP32338.1 GBBM01002753 JAC32665.1 JO843142 AEO34759.1 GBBK01003929 JAC20553.1 GBBL01002061 JAC25259.1 GFAC01006942 JAT92246.1 GFJQ02008288 JAV98681.1 KQ971347 EFA05502.1 GAMD01002801 JAA98789.1 GGFM01004129 MBW24880.1 GGFK01009131 MBW42452.1 ADMH02001603 GGFL01004223 ETN61873.1 MBW68401.1 GFWV01005447 MAA30177.1 KK121265 KFM80092.1 BT128085 AEE63046.1 ABJB010872459 DS615069 EEC00336.1 GADI01001594 GEFM01003767 JAA72214.1 JAP72029.1 AXCM01003145 AXCN02000047 AL844518 BC045981 BC071331 HADZ01020103 SBP84044.1 HADY01020028 SBP58513.1 HAEH01014538 SBS00351.1 AAAB01008859 EAA08218.2 APCN01000459 BDOT01000138 GCF52634.1 GGFK01009608 MBW42929.1 GBYX01137196 JAO94622.1 CM012447 RVE66327.1 KK854881 PTY19444.1 ATLV01012143 KE524764 KFB36212.1 GDKW01000775 JAI55820.1 ACPB03015836 GAHY01000775 JAA76735.1 KQ978023 KYM97961.1 QUSF01000030 RLV99829.1 MUZQ01000045 OWK61161.1 QRBI01000121 RMC05787.1 AWGT02000001 OXB84910.1 LSYS01006629 OPJ75016.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000078540
+ More
UP000075809 UP000053097 UP000279307 UP000053825 UP000242457 UP000078541 UP000053105 UP000192223 UP000005205 UP000007755 UP000002358 UP000027135 UP000007266 UP000000673 UP000069272 UP000054359 UP000001555 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000075881 UP000075920 UP000000437 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000192220 UP000288954 UP000264800 UP000261640 UP000075880 UP000265000 UP000261480 UP000002852 UP000261560 UP000261500 UP000242638 UP000001038 UP000265180 UP000265200 UP000030765 UP000015103 UP000078542 UP000276834 UP000197619 UP000269221 UP000198419 UP000190648
UP000075809 UP000053097 UP000279307 UP000053825 UP000242457 UP000078541 UP000053105 UP000192223 UP000005205 UP000007755 UP000002358 UP000027135 UP000007266 UP000000673 UP000069272 UP000054359 UP000001555 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000075881 UP000075920 UP000000437 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000192220 UP000288954 UP000264800 UP000261640 UP000075880 UP000265000 UP000261480 UP000002852 UP000261560 UP000261500 UP000242638 UP000001038 UP000265180 UP000265200 UP000030765 UP000015103 UP000078542 UP000276834 UP000197619 UP000269221 UP000198419 UP000190648
Pfam
PF12850 Metallophos_2
Interpro
Gene 3D
ProteinModelPortal
H9JUH7
Q1HPG7
I4DQC7
A0A3S2P9Y1
A0A2A4J009
S4PJX1
+ More
A0A212F235 A0A0N1PIZ8 A0A195BVK4 A0A151WSX7 A0A026VWC9 A0A0L7QPQ8 A0A2A3E7G3 A0A195FQC0 A0A0M8ZUJ4 A0A1W4XM03 A0A1S4N1Z2 A0A158NV20 F4WXI8 A0A1B6KBG3 A0A1B6IRK5 A0A1B6EIF4 R4V4Z7 A0A1B6EA34 A0A224YT62 A0A131YGQ7 L7M701 A0A131XBB0 K7ITM8 A0A1A7WMD7 A0A067RKM6 A0A1A8D8K1 A0A1A8KNE5 A0A1A8UC64 A0A1A8SDI7 A0A1A8Q0G0 A0A1Y1KZP6 A0A2R5LLT0 A0A1A7YQ83 A0A023GIR9 G3MMP1 A0A023FIU7 A0A023FUV0 A0A1E1WZN2 A0A1Z5KUA4 D2A6E5 T1DGE8 A0A2M3Z8R8 A0A2M4ANY8 W5JEV3 A0A182FR85 A0A293LA52 A0A087URV3 J3JXW3 B7P164 A0A0K8RNP5 A0A182LTS8 A0A182RNL6 A0A182N4L6 A0A182PZU0 A0A182Y7B1 A0A182K5M4 A0A182VQ81 Q7ZV68 A0A1A8CWC6 A0A1A8AV03 A0A1A8R307 A0A182PTH3 A0A182VHK4 A0A182TUD7 A0A182XEI3 A0A182LFI1 Q7QBW4 A0A182HTC9 A0A2I4CC29 A0A402F869 A0A3Q3B3M9 A0A2M4AQ85 A0A3Q3KSW7 A0A182IZA4 A0A3Q2P9J9 A0A3B3Y244 A0A0S7LYW6 A0A3B5RA41 A0A3B3D2V0 A0A3B3UAZ5 A0A3P9NFP8 H2MUI2 A0A3S2PPD7 A0A3P9KV97 A0A2R7WIG8 A0A084VE18 A0A0P4VUV8 R4G562 A0A195CCK9 A0A3L8SD62 A0A218V657 A0A3M0KF96 A0A226PZG7 A0A1V4JS14
A0A212F235 A0A0N1PIZ8 A0A195BVK4 A0A151WSX7 A0A026VWC9 A0A0L7QPQ8 A0A2A3E7G3 A0A195FQC0 A0A0M8ZUJ4 A0A1W4XM03 A0A1S4N1Z2 A0A158NV20 F4WXI8 A0A1B6KBG3 A0A1B6IRK5 A0A1B6EIF4 R4V4Z7 A0A1B6EA34 A0A224YT62 A0A131YGQ7 L7M701 A0A131XBB0 K7ITM8 A0A1A7WMD7 A0A067RKM6 A0A1A8D8K1 A0A1A8KNE5 A0A1A8UC64 A0A1A8SDI7 A0A1A8Q0G0 A0A1Y1KZP6 A0A2R5LLT0 A0A1A7YQ83 A0A023GIR9 G3MMP1 A0A023FIU7 A0A023FUV0 A0A1E1WZN2 A0A1Z5KUA4 D2A6E5 T1DGE8 A0A2M3Z8R8 A0A2M4ANY8 W5JEV3 A0A182FR85 A0A293LA52 A0A087URV3 J3JXW3 B7P164 A0A0K8RNP5 A0A182LTS8 A0A182RNL6 A0A182N4L6 A0A182PZU0 A0A182Y7B1 A0A182K5M4 A0A182VQ81 Q7ZV68 A0A1A8CWC6 A0A1A8AV03 A0A1A8R307 A0A182PTH3 A0A182VHK4 A0A182TUD7 A0A182XEI3 A0A182LFI1 Q7QBW4 A0A182HTC9 A0A2I4CC29 A0A402F869 A0A3Q3B3M9 A0A2M4AQ85 A0A3Q3KSW7 A0A182IZA4 A0A3Q2P9J9 A0A3B3Y244 A0A0S7LYW6 A0A3B5RA41 A0A3B3D2V0 A0A3B3UAZ5 A0A3P9NFP8 H2MUI2 A0A3S2PPD7 A0A3P9KV97 A0A2R7WIG8 A0A084VE18 A0A0P4VUV8 R4G562 A0A195CCK9 A0A3L8SD62 A0A218V657 A0A3M0KF96 A0A226PZG7 A0A1V4JS14
PDB
3PSO
E-value=7.12811e-91,
Score=847
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Membrane
Endosome membrane
Membrane
Endosome membrane
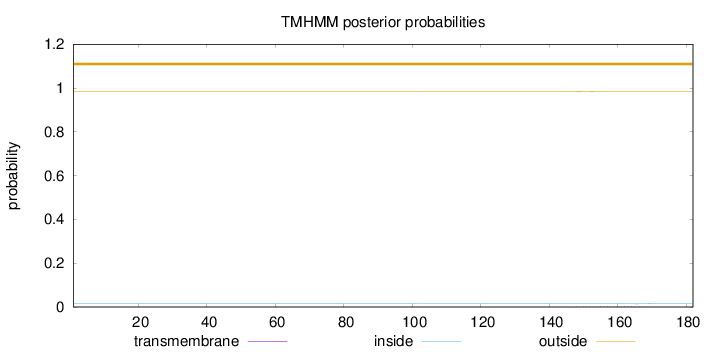
Length:
182
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03472
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.01445
outside
1 - 182
Population Genetic Test Statistics
Pi
294.235536
Theta
183.127174
Tajima's D
1.852285
CLR
0.078346
CSRT
0.857207139643018
Interpretation
Uncertain
