Pre Gene Modal
BGIBMGA013232
Annotation
PREDICTED:_peptidyl-tRNA_hydrolase_2?_mitochondrial-like_[Papilio_xuthus]
Full name
Peptidyl-tRNA hydrolase 2, mitochondrial
Alternative Name
Bcl-2 inhibitor of transcription
Bcl-2 inhibitor of transcription 1
Bcl-2 inhibitor of transcription 1
Location in the cell
Cytoplasmic Reliability : 1.778 Mitochondrial Reliability : 1.327
Sequence
CDS
ATGACGTTCGATATGTCTTTTTACACTGGCCTAAGCTGCGGTATTTGTATCGGCCTATCAATATTTGCGATTCGAAAGTACATGAAGATGTTTTCTAATGCCAGTGAAACTGTTAAAAAGTTTGTATCGAATGAAGAGTACAAATTGGTTCTAGTTGTCCGAACAGACCTAAGTATGGGTAAAGGCAAAATAGCAGCTCAATGCTGTCATGCAGCAGTTGGAGCTTTTGAGAAAGCACTAAAAAAAGACCCTGAAGGTTTGAAAGCCTGGCAAATGACAGGACAGGCTAAAGTGGCCCTTAAAATTGACTCTTTAGAGGAAATAAAAAAAATTGCAGACAATGCAAAGAAAATGGGCTTAATCACATCTTTAATACGTGATGCGGGCCGTACCCAGATTGCACCAAATTCAATAACAGTTTTGGGTGTAGGACCAGCACCGAAAGACATTATAGACAAAGTCACCGGTCACCTTAAGTTGCTGTGA
Protein
MTFDMSFYTGLSCGICIGLSIFAIRKYMKMFSNASETVKKFVSNEEYKLVLVVRTDLSMGKGKIAAQCCHAAVGAFEKALKKDPEGLKAWQMTGQAKVALKIDSLEEIKKIADNAKKMGLITSLIRDAGRTQIAPNSITVLGVGPAPKDIIDKVTGHLKLL
Summary
Description
The natural substrate for this enzyme may be peptidyl-tRNAs which drop off the ribosome during protein synthesis.
Promotes caspase-independent apoptosis by regulating the function of two transcriptional regulators, AES and TLE1.
Promotes caspase-independent apoptosis by regulating the function of two transcriptional regulators, AES and TLE1.
Catalytic Activity
H2O + N-acyl-L-alpha-aminoacyl-tRNA = an N-acyl-L-amino acid + H(+) + tRNA
Subunit
Monomer.
Similarity
Belongs to the PTH2 family.
Keywords
Complete proteome
Hydrolase
Isopeptide bond
Mitochondrion
Reference proteome
Transit peptide
Ubl conjugation
3D-structure
Apoptosis
Disease mutation
Feature
chain Peptidyl-tRNA hydrolase 2, mitochondrial
sequence variant In IMNEPD; dbSNP:rs730882234.
sequence variant In IMNEPD; dbSNP:rs730882234.
Uniprot
H9JUL9
A0A2A4JPI5
A0A437B4X6
A0A2H1V2R5
A0A0N0PC43
A0A212FJ33
+ More
A0A1E1WPF4 A0A0L7K3A7 A4IGX7 A0A2K6SMI2 A0A1B0C8E5 A0A091PCS3 A0A2T7PD38 A0A2K5CE31 A0A1L8HFU3 A0A3Q0EI15 A0A1U7UK08 F7HZT2 U3EMU8 A0A091VSR0 F6PQA8 A0A2R5L7F2 A0A2K6ETW0 A0A1S4FCF4 A0A2D4BRB5 A0A2K5PJ50 E1BWS4 A0A023EHQ7 A0A0D9S9B4 E2RND8 H0XIT5 A0A452E2A9 W5P154 K3WU49 A0A2Y9I750 A0A3Q7S7E7 Q177N9 F1S227 A0A2U3VR79 G1NR67 M7BPU4 A0A226PG38 A0A2U3Y9I0 A0A3Q7X793 A0A384CGZ8 A0A3Q7MK85 A0A091IUC8 A0A226NBN1 W5LT50 A0A091SX53 A0A452R180 A0A452R182 A0A3Q7XFP4 A0A384CFX6 A0A286XAG6 Q3ZBL5 L8IR93 A0A094KE74 G3UA53 E9H345 A0A067R5D2 A0A093R4V7 A0A2P4XPH0 A0A2K6L8S4 A0A2K6QIN2 A0A2K5JGL3 A0A1Z5LD88 A0A2Y9JWZ1 M3WNP1 Q9NAZ7 G7PUH4 F7GV88 A0A2K5TM78 A0A2K5XY13 A0A2K5NV60 A0A2K6CUM5 A0A096P4W9 A0A1D5QQ67 A0A340WQ00 A0A091PL18 Q96ME4 A0A091NBE6 G1QJ28 A0A2J8W7L2 A0A093NGX2 A0A2J8W7J7 D2HLU6 G9L1D3 A0A091FV65 Q7QK06 A0A3P4NL33 M3Z7D7 U6CTF4 A0A293LIY7 K7DQ09 Q9Y3E5 A0A2R9AL47 H2QDK5 J3KQ48
A0A1E1WPF4 A0A0L7K3A7 A4IGX7 A0A2K6SMI2 A0A1B0C8E5 A0A091PCS3 A0A2T7PD38 A0A2K5CE31 A0A1L8HFU3 A0A3Q0EI15 A0A1U7UK08 F7HZT2 U3EMU8 A0A091VSR0 F6PQA8 A0A2R5L7F2 A0A2K6ETW0 A0A1S4FCF4 A0A2D4BRB5 A0A2K5PJ50 E1BWS4 A0A023EHQ7 A0A0D9S9B4 E2RND8 H0XIT5 A0A452E2A9 W5P154 K3WU49 A0A2Y9I750 A0A3Q7S7E7 Q177N9 F1S227 A0A2U3VR79 G1NR67 M7BPU4 A0A226PG38 A0A2U3Y9I0 A0A3Q7X793 A0A384CGZ8 A0A3Q7MK85 A0A091IUC8 A0A226NBN1 W5LT50 A0A091SX53 A0A452R180 A0A452R182 A0A3Q7XFP4 A0A384CFX6 A0A286XAG6 Q3ZBL5 L8IR93 A0A094KE74 G3UA53 E9H345 A0A067R5D2 A0A093R4V7 A0A2P4XPH0 A0A2K6L8S4 A0A2K6QIN2 A0A2K5JGL3 A0A1Z5LD88 A0A2Y9JWZ1 M3WNP1 Q9NAZ7 G7PUH4 F7GV88 A0A2K5TM78 A0A2K5XY13 A0A2K5NV60 A0A2K6CUM5 A0A096P4W9 A0A1D5QQ67 A0A340WQ00 A0A091PL18 Q96ME4 A0A091NBE6 G1QJ28 A0A2J8W7L2 A0A093NGX2 A0A2J8W7J7 D2HLU6 G9L1D3 A0A091FV65 Q7QK06 A0A3P4NL33 M3Z7D7 U6CTF4 A0A293LIY7 K7DQ09 Q9Y3E5 A0A2R9AL47 H2QDK5 J3KQ48
EC Number
3.1.1.29
Pubmed
19121390
26354079
22118469
26227816
27762356
25243066
+ More
20431018 15592404 24945155 26483478 16341006 20809919 17510324 30723633 20838655 23624526 24621616 24813606 25329095 21993624 16305752 22751099 21292972 24845553 28186564 25362486 28528879 17975172 11762196 22002653 17431167 25319552 14702039 20010809 23236062 12364791 14747013 17210077 10810093 15489334 17974005 15006356 18669648 21269460 23186163 25574476 25558065 25621951 25944712 14660562 22722832 16136131 16625196
20431018 15592404 24945155 26483478 16341006 20809919 17510324 30723633 20838655 23624526 24621616 24813606 25329095 21993624 16305752 22751099 21292972 24845553 28186564 25362486 28528879 17975172 11762196 22002653 17431167 25319552 14702039 20010809 23236062 12364791 14747013 17210077 10810093 15489334 17974005 15006356 18669648 21269460 23186163 25574476 25558065 25621951 25944712 14660562 22722832 16136131 16625196
EMBL
BABH01022680
NWSH01000975
PCG73320.1
RSAL01000158
RVE45541.1
ODYU01000380
+ More
SOQ35066.1 KQ460655 KPJ12954.1 AGBW02008320 OWR53720.1 GDQN01002182 JAT88872.1 JTDY01013301 KOB52159.1 BC135286 AAI35287.1 AJWK01000518 KL384128 KFP89320.1 PZQS01000004 PVD31332.1 CM004468 OCT94962.1 GAMS01006065 GAMR01010587 GAMQ01000451 GAMP01006873 JAB17071.1 JAB23345.1 JAB41400.1 JAB45882.1 KK734272 KFR05518.1 AAMC01027133 GGLE01001266 MBY05392.1 BBXB01000050 GAX96580.1 AADN05000057 JXUM01127242 JXUM01132779 GAPW01005021 KQ567891 KQ567165 JAC08577.1 KXJ69236.1 KXJ69585.1 AQIB01117261 AAEX03006586 AAQR03121548 LWLT01000022 AMGL01017392 GL376613 CH477373 EAT42379.1 AEMK02000080 DQIR01116625 DQIR01254069 HDA72101.1 HDC09547.1 KB521250 EMP37730.1 AWGT02000074 OXB79045.1 KK501002 KFP11912.1 MCFN01000109 OXB64898.1 KK936918 KFQ47675.1 AAKN02033032 AB238942 BT021191 BC103230 JH880797 ELR58836.1 KL344832 KFZ55168.1 GL732587 EFX73896.1 KK852685 KDR18489.1 KL438923 KFW93645.1 NCKW01009159 POM67434.1 GFJQ02002059 JAW04911.1 AANG04003358 AF275673 AAF87580.1 CM001291 EHH58023.1 JSUE03017039 JU475959 JV047344 CM001268 AFH32763.1 AFI37415.1 EHH24869.1 AQIA01027655 AHZZ02010224 KK674812 KFQ07978.1 AK057033 BAB71351.2 KK828104 KFP74329.1 ADFV01107753 NDHI03003397 PNJ65731.1 KL224565 KFW61245.1 ABGA01152269 PNJ65732.1 PNJ65733.1 GL193022 EFB14817.1 JP022364 AES10962.1 KL447514 KFO74405.1 AAAB01008805 EAA03889.5 CYRY02026482 VCW98752.1 AEYP01021737 HAAF01001945 CCP73771.1 GFWV01003953 MAA28683.1 GABC01009269 GABF01006306 GABD01006927 GABE01006322 NBAG03000258 JAA02069.1 JAA15839.1 JAA26173.1 JAA38417.1 PNI58065.1 PNI58066.1 AF151905 AK098219 CH471109 BC006807 AL137322 AJFE02058761 AACZ04026964 PNI58064.1 AC091271
SOQ35066.1 KQ460655 KPJ12954.1 AGBW02008320 OWR53720.1 GDQN01002182 JAT88872.1 JTDY01013301 KOB52159.1 BC135286 AAI35287.1 AJWK01000518 KL384128 KFP89320.1 PZQS01000004 PVD31332.1 CM004468 OCT94962.1 GAMS01006065 GAMR01010587 GAMQ01000451 GAMP01006873 JAB17071.1 JAB23345.1 JAB41400.1 JAB45882.1 KK734272 KFR05518.1 AAMC01027133 GGLE01001266 MBY05392.1 BBXB01000050 GAX96580.1 AADN05000057 JXUM01127242 JXUM01132779 GAPW01005021 KQ567891 KQ567165 JAC08577.1 KXJ69236.1 KXJ69585.1 AQIB01117261 AAEX03006586 AAQR03121548 LWLT01000022 AMGL01017392 GL376613 CH477373 EAT42379.1 AEMK02000080 DQIR01116625 DQIR01254069 HDA72101.1 HDC09547.1 KB521250 EMP37730.1 AWGT02000074 OXB79045.1 KK501002 KFP11912.1 MCFN01000109 OXB64898.1 KK936918 KFQ47675.1 AAKN02033032 AB238942 BT021191 BC103230 JH880797 ELR58836.1 KL344832 KFZ55168.1 GL732587 EFX73896.1 KK852685 KDR18489.1 KL438923 KFW93645.1 NCKW01009159 POM67434.1 GFJQ02002059 JAW04911.1 AANG04003358 AF275673 AAF87580.1 CM001291 EHH58023.1 JSUE03017039 JU475959 JV047344 CM001268 AFH32763.1 AFI37415.1 EHH24869.1 AQIA01027655 AHZZ02010224 KK674812 KFQ07978.1 AK057033 BAB71351.2 KK828104 KFP74329.1 ADFV01107753 NDHI03003397 PNJ65731.1 KL224565 KFW61245.1 ABGA01152269 PNJ65732.1 PNJ65733.1 GL193022 EFB14817.1 JP022364 AES10962.1 KL447514 KFO74405.1 AAAB01008805 EAA03889.5 CYRY02026482 VCW98752.1 AEYP01021737 HAAF01001945 CCP73771.1 GFWV01003953 MAA28683.1 GABC01009269 GABF01006306 GABD01006927 GABE01006322 NBAG03000258 JAA02069.1 JAA15839.1 JAA26173.1 JAA38417.1 PNI58065.1 PNI58066.1 AF151905 AK098219 CH471109 BC006807 AL137322 AJFE02058761 AACZ04026964 PNI58064.1 AC091271
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000037510
+ More
UP000233220 UP000092461 UP000245119 UP000233020 UP000186698 UP000189704 UP000008225 UP000053605 UP000008143 UP000233160 UP000054177 UP000233040 UP000000539 UP000069940 UP000249989 UP000029965 UP000002254 UP000005225 UP000291000 UP000002356 UP000248481 UP000286640 UP000008820 UP000008227 UP000245340 UP000001645 UP000031443 UP000198419 UP000245341 UP000286642 UP000261680 UP000291021 UP000286641 UP000053119 UP000198323 UP000018467 UP000291022 UP000005447 UP000009136 UP000007646 UP000000305 UP000027135 UP000233180 UP000233200 UP000233080 UP000248482 UP000011712 UP000009130 UP000006718 UP000233100 UP000233140 UP000233060 UP000233120 UP000028761 UP000265300 UP000001073 UP000054081 UP000001595 UP000053760 UP000007062 UP000000715 UP000005640 UP000240080 UP000002277
UP000233220 UP000092461 UP000245119 UP000233020 UP000186698 UP000189704 UP000008225 UP000053605 UP000008143 UP000233160 UP000054177 UP000233040 UP000000539 UP000069940 UP000249989 UP000029965 UP000002254 UP000005225 UP000291000 UP000002356 UP000248481 UP000286640 UP000008820 UP000008227 UP000245340 UP000001645 UP000031443 UP000198419 UP000245341 UP000286642 UP000261680 UP000291021 UP000286641 UP000053119 UP000198323 UP000018467 UP000291022 UP000005447 UP000009136 UP000007646 UP000000305 UP000027135 UP000233180 UP000233200 UP000233080 UP000248482 UP000011712 UP000009130 UP000006718 UP000233100 UP000233140 UP000233060 UP000233120 UP000028761 UP000265300 UP000001073 UP000054081 UP000001595 UP000053760 UP000007062 UP000000715 UP000005640 UP000240080 UP000002277
Pfam
PF01981 PTH2
SUPFAM
SSF102462
SSF102462
Gene 3D
CDD
ProteinModelPortal
H9JUL9
A0A2A4JPI5
A0A437B4X6
A0A2H1V2R5
A0A0N0PC43
A0A212FJ33
+ More
A0A1E1WPF4 A0A0L7K3A7 A4IGX7 A0A2K6SMI2 A0A1B0C8E5 A0A091PCS3 A0A2T7PD38 A0A2K5CE31 A0A1L8HFU3 A0A3Q0EI15 A0A1U7UK08 F7HZT2 U3EMU8 A0A091VSR0 F6PQA8 A0A2R5L7F2 A0A2K6ETW0 A0A1S4FCF4 A0A2D4BRB5 A0A2K5PJ50 E1BWS4 A0A023EHQ7 A0A0D9S9B4 E2RND8 H0XIT5 A0A452E2A9 W5P154 K3WU49 A0A2Y9I750 A0A3Q7S7E7 Q177N9 F1S227 A0A2U3VR79 G1NR67 M7BPU4 A0A226PG38 A0A2U3Y9I0 A0A3Q7X793 A0A384CGZ8 A0A3Q7MK85 A0A091IUC8 A0A226NBN1 W5LT50 A0A091SX53 A0A452R180 A0A452R182 A0A3Q7XFP4 A0A384CFX6 A0A286XAG6 Q3ZBL5 L8IR93 A0A094KE74 G3UA53 E9H345 A0A067R5D2 A0A093R4V7 A0A2P4XPH0 A0A2K6L8S4 A0A2K6QIN2 A0A2K5JGL3 A0A1Z5LD88 A0A2Y9JWZ1 M3WNP1 Q9NAZ7 G7PUH4 F7GV88 A0A2K5TM78 A0A2K5XY13 A0A2K5NV60 A0A2K6CUM5 A0A096P4W9 A0A1D5QQ67 A0A340WQ00 A0A091PL18 Q96ME4 A0A091NBE6 G1QJ28 A0A2J8W7L2 A0A093NGX2 A0A2J8W7J7 D2HLU6 G9L1D3 A0A091FV65 Q7QK06 A0A3P4NL33 M3Z7D7 U6CTF4 A0A293LIY7 K7DQ09 Q9Y3E5 A0A2R9AL47 H2QDK5 J3KQ48
A0A1E1WPF4 A0A0L7K3A7 A4IGX7 A0A2K6SMI2 A0A1B0C8E5 A0A091PCS3 A0A2T7PD38 A0A2K5CE31 A0A1L8HFU3 A0A3Q0EI15 A0A1U7UK08 F7HZT2 U3EMU8 A0A091VSR0 F6PQA8 A0A2R5L7F2 A0A2K6ETW0 A0A1S4FCF4 A0A2D4BRB5 A0A2K5PJ50 E1BWS4 A0A023EHQ7 A0A0D9S9B4 E2RND8 H0XIT5 A0A452E2A9 W5P154 K3WU49 A0A2Y9I750 A0A3Q7S7E7 Q177N9 F1S227 A0A2U3VR79 G1NR67 M7BPU4 A0A226PG38 A0A2U3Y9I0 A0A3Q7X793 A0A384CGZ8 A0A3Q7MK85 A0A091IUC8 A0A226NBN1 W5LT50 A0A091SX53 A0A452R180 A0A452R182 A0A3Q7XFP4 A0A384CFX6 A0A286XAG6 Q3ZBL5 L8IR93 A0A094KE74 G3UA53 E9H345 A0A067R5D2 A0A093R4V7 A0A2P4XPH0 A0A2K6L8S4 A0A2K6QIN2 A0A2K5JGL3 A0A1Z5LD88 A0A2Y9JWZ1 M3WNP1 Q9NAZ7 G7PUH4 F7GV88 A0A2K5TM78 A0A2K5XY13 A0A2K5NV60 A0A2K6CUM5 A0A096P4W9 A0A1D5QQ67 A0A340WQ00 A0A091PL18 Q96ME4 A0A091NBE6 G1QJ28 A0A2J8W7L2 A0A093NGX2 A0A2J8W7J7 D2HLU6 G9L1D3 A0A091FV65 Q7QK06 A0A3P4NL33 M3Z7D7 U6CTF4 A0A293LIY7 K7DQ09 Q9Y3E5 A0A2R9AL47 H2QDK5 J3KQ48
PDB
1Q7S
E-value=3.22859e-34,
Score=357
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
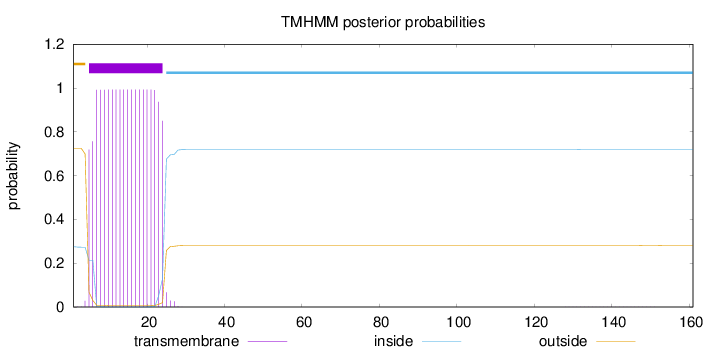
Length:
161
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.31256
Exp number, first 60 AAs:
19.30447
Total prob of N-in:
0.27538
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 24
inside
25 - 161
Population Genetic Test Statistics
Pi
157.617782
Theta
53.705453
Tajima's D
1.395659
CLR
1.309748
CSRT
0.76301184940753
Interpretation
Uncertain
