Gene
KWMTBOMO09952 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013237
Annotation
cytochrome_P450_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.554
Sequence
CDS
ATGTCAGTGAGCGCATTAGTATTTTCCGCCTTCGTTTTATTAGTGACGTATATTTACTATTGGTCGACCCGGAAATTCGATTATTGGAAGCGAAAAAATGTGCCATACGCTAAACCTGTGCCGTTCTTTGGGAACTACATGCGATATATCACACTGCAGAGCTTCCTAGGAGATGTGATGCAGAAGCTGTGCCAGCAGTTCCCTGACAGACCGTACTTCGGATCGTTTTACGGTACGGAACCAGCCCTAGTCTTGCAGAATCCGGAGATCATCAAGCAAGTTTTTACTAAAGACTTCTATTACTTCAACAGCCGCGAAAACAGGGATTACAATCATAAGGAAGTATTCACACAGAACTTGTTCTTCGCCAATGGAGATAGATGGAAGGTAATACGTCAGAACCTAACCCCGCTGTTCTCATCGTCAAAGATGAAGAATATGTTTTACTTGGTGGAAAAATGCAACCATTCCCTTGAAGACATGCTGGACAAGGAAACCAAAGACTTGCAAAGTATTGAGATCCGGTCTGCTATGATAAGGTACACACTGGACTCTATCTGCTCTAGCGCCTTCGGTATCGAAACAAATACCTTAAGTGAGGGCGCAGAAAATAGTCCTTTTCCAAGCATGGGAAGCACCATATTTAGTTCTTCGATAACTCGAGGACTGAAATTGATTGGTCGTTCGATGTGGCCGGGCATATTCTATAAGCTTGGTCTCCGGTGCTTTCCAACGGAAATTGACGATTTCTTTGAGAGACTCCTGACTGAGGTGTTCGAGAACCGTGGCTACAAGCCGACGAACCGTAACGATTTCGTGGACTTAATCTTGAGTCTGAAACAAAACGATTACCTGACCGGAGATGGCTTGGTTCCCAAAAACGTGGACGCAAAGAAAGTGACGGTCAAGGTTGACGACGCCTTATTGATAGCGCAGTGCGTAGTCTTCTTTGGTGCAGGCTTCGAAACGTCAGCTACCACTCTCTCGGCAGCCCTATACGAGCTGGCCAAGAACCCAGAAGCCCAAAGGCGCGCTCAAGAGGAAGTTGACGAACTTTTGCTAAAACACAACAACAAGTTAAATTATGATTGTTTAGCAGAATTACCGTACTTGGAGGCATGTATGAACGAAGCGATGCGTCTGTACCCAGTATTTCCTAATATCACCCGCGAAACCGTTGCCGACTATACGTTTCCCGATGGCCTAAGGATAGATAAGGGTATGCGTGTGCATGTCCCGGTGTATGCTATCCACCGTAACCCCGACAACTTTCCTGACCCGGAAGAGTTTCGTCCTGAGAGACATTTAGGAGACGCCAAAAACGACATTAAGCAGTTCACGTTCTTCCCGTTCGGAGAAGGGCCCAGGATATGCATTGGTATGCGCTTCGGGAAAATGCAGACTATCGCTGGGCTGATTACCTGTTTGAAAAAGTTCAACTTCGAACTGGCGGACGGTATGCCGAGGACGCTAGCTTTTAGATCCACAACGTTGCTTACACAGCCGAGTACCGGATTATTCCTGAAGGCCACACCACGAGACGGATGGGAGCAACGTATTTTCGCACGTTAA
Protein
MSVSALVFSAFVLLVTYIYYWSTRKFDYWKRKNVPYAKPVPFFGNYMRYITLQSFLGDVMQKLCQQFPDRPYFGSFYGTEPALVLQNPEIIKQVFTKDFYYFNSRENRDYNHKEVFTQNLFFANGDRWKVIRQNLTPLFSSSKMKNMFYLVEKCNHSLEDMLDKETKDLQSIEIRSAMIRYTLDSICSSAFGIETNTLSEGAENSPFPSMGSTIFSSSITRGLKLIGRSMWPGIFYKLGLRCFPTEIDDFFERLLTEVFENRGYKPTNRNDFVDLILSLKQNDYLTGDGLVPKNVDAKKVTVKVDDALLIAQCVVFFGAGFETSATTLSAALYELAKNPEAQRRAQEEVDELLLKHNNKLNYDCLAELPYLEACMNEAMRLYPVFPNITRETVADYTFPDGLRIDKGMRVHVPVYAIHRNPDNFPDPEEFRPERHLGDAKNDIKQFTFFPFGEGPRICIGMRFGKMQTIAGLITCLKKFNFELADGMPRTLAFRSTTLLTQPSTGLFLKATPRDGWEQRIFAR
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N766
L0N7C5
D6MLU9
B6VFR9
A9YXV8
A4GUB9
+ More
A5JM34 A9QW13 A5HG34 H9JUM4 A3RIC0 L0N6H5 A0A172MF86 D5L0M6 A0A0L7KJY7 A0A248QEX7 A0A0L7LJH7 A0A2A4J698 A0A068EWQ5 Q2XSW1 A0A068EU72 A0A068EVK6 A0A068F0X2 A0A0L7LRY0 A0A068EVD2 A0A2A4J2E3 A0A068EVL0 Q7YZS2 A0A0K8TUL3 A0A068ETU4 H2CZF1 H2CZE9 A0A0L7KVL1 A0A068EU65 F4YCW0 A0A3S2LFF7 A0A0K0YD30 Q069I9 A0A2A4JAM9 A0A2A4JAK3 A0A068ETU8 A0A248QHF4 A0A2A4J7V4 B3VT94 A0A248QEF8 A0A068F1Z7 A0A2H1V0E7 A0A2H1WID4 A0A2A4J918 J7FKN2 A0A0C5C1I6 A0A0K8TUI1 A0A3S2P9Y7 C1LZ54 J7FJF2 H9JUM8 A4GUB8 A0A437B5H6 A0A1V0D9E0 A0A1V0D9D9 A0A0N1IP03 A0A1V0D9D6 J7FKN1 A0A068EWQ0 X5D9B5 A0A411E5Q0 A0A0A7DH57 A0A2H1V048 A0A0N1ID06 A0A286QUG7 D2JLK6 W5VY13 A9QW15 A0A3S2LW02 A5HKM1 A0A0C5CGV6 A0A2A4JWC6 A0A222NX20 A0A0K8TV92 A0A2W1BGY9 L0N6M3 C7U1L4 A0A068F0X7 A0A248QG18 A0A0A7DH10 H9JUM5 J7FJF3 A0A2H1VU17 A0A248QEH8 A0A0A7DGX3 A0A0A7DH65 A5HG33 H9JUM6 A0A411AG09 A0A0A7DG86 D5L0M5 A0A0A7DH14 A0A2H1VSH4 A0A2H1VSG7
A5JM34 A9QW13 A5HG34 H9JUM4 A3RIC0 L0N6H5 A0A172MF86 D5L0M6 A0A0L7KJY7 A0A248QEX7 A0A0L7LJH7 A0A2A4J698 A0A068EWQ5 Q2XSW1 A0A068EU72 A0A068EVK6 A0A068F0X2 A0A0L7LRY0 A0A068EVD2 A0A2A4J2E3 A0A068EVL0 Q7YZS2 A0A0K8TUL3 A0A068ETU4 H2CZF1 H2CZE9 A0A0L7KVL1 A0A068EU65 F4YCW0 A0A3S2LFF7 A0A0K0YD30 Q069I9 A0A2A4JAM9 A0A2A4JAK3 A0A068ETU8 A0A248QHF4 A0A2A4J7V4 B3VT94 A0A248QEF8 A0A068F1Z7 A0A2H1V0E7 A0A2H1WID4 A0A2A4J918 J7FKN2 A0A0C5C1I6 A0A0K8TUI1 A0A3S2P9Y7 C1LZ54 J7FJF2 H9JUM8 A4GUB8 A0A437B5H6 A0A1V0D9E0 A0A1V0D9D9 A0A0N1IP03 A0A1V0D9D6 J7FKN1 A0A068EWQ0 X5D9B5 A0A411E5Q0 A0A0A7DH57 A0A2H1V048 A0A0N1ID06 A0A286QUG7 D2JLK6 W5VY13 A9QW15 A0A3S2LW02 A5HKM1 A0A0C5CGV6 A0A2A4JWC6 A0A222NX20 A0A0K8TV92 A0A2W1BGY9 L0N6M3 C7U1L4 A0A068F0X7 A0A248QG18 A0A0A7DH10 H9JUM5 J7FJF3 A0A2H1VU17 A0A248QEH8 A0A0A7DGX3 A0A0A7DH65 A5HG33 H9JUM6 A0A411AG09 A0A0A7DG86 D5L0M5 A0A0A7DH14 A0A2H1VSH4 A0A2H1VSG7
Pubmed
EMBL
AK343205
BAM73891.1
AK289291
AK343204
BAM73818.1
BAM73890.1
+ More
GQ241737 ADD70250.1 FJ387169 ACJ10074.1 EU309477 ABY40426.1 EF432657 ABO21683.1 EF547545 ABQ42339.1 EU273875 ABX64438.1 EF528486 ABP99020.1 BABH01022694 EF415296 ABN71367.1 AK289290 BAM73817.1 KU145393 ANC96676.1 GU731531 ADE05581.1 JTDY01009617 KOB63214.1 KX443435 ASO98010.1 JTDY01000942 KOB75351.1 NWSH01002736 PCG67605.1 KM016744 AID54896.1 DQ256407 ABB69054.1 KM016742 AID54894.1 KM016736 AID54888.1 KM016738 AID54890.1 JTDY01000272 KOB77971.1 KJ671575 AID55427.1 NWSH01003425 PCG66327.1 KM016741 AID54893.1 AY295774 AAP83689.1 GCVX01000058 JAI18172.1 KM016735 AID54887.1 JN375491 AEY75585.1 JN375489 AEY75583.1 JTDY01005369 KOB67074.1 KM016737 AID54889.1 GU937509 ADW23116.1 RSAL01000158 RVE45547.1 KR095600 AKS48888.1 DQ986461 JN176574 ABI84381.1 AET11931.1 NWSH01002369 PCG68453.1 PCG68452.1 KM016740 AID54892.1 KX443432 ASO98007.1 PCG67604.1 EU807990 ACF17813.2 KX443430 ASO98005.1 KJ671576 AID55428.1 ODYU01000082 SOQ34236.1 ODYU01008863 SOQ52829.1 PCG68451.1 JX310082 AFP20593.1 KP001126 AJN91171.1 GCVX01000081 JAI18149.1 RVE45548.1 FN356971 CAX94849.1 JX310078 AFP20589.1 BABH01022705 EF432656 ABO21682.1 RVE45549.1 KY212044 ARA91608.1 KY212043 ARA91607.1 KQ460655 KPJ12961.1 KY212045 ARA91609.1 JX310077 AFP20588.1 KM016739 AID54891.1 KF701137 AHW57307.1 MH236461 QBA57466.1 KJ645906 AIJ00763.1 ODYU01000083 SOQ34238.1 KQ459388 KPJ01135.1 KX443431 ASO98006.1 GQ915322 ACZ97416.2 KF853191 AHH92929.1 EU273877 ABX64440.1 RVE45546.1 EF535809 ABQ08711.1 KP001127 AJN91172.1 NWSH01000502 PCG75998.1 KX844829 ASQ44127.1 GCVX01000049 JAI18181.1 KZ150141 PZC72944.1 AK343206 BAM73892.1 FN544262 CBB07053.1 KM016743 AID54895.1 KX443433 ASO98008.1 KJ645904 AIJ00761.1 JX310083 AFP20594.1 ODYU01004424 SOQ44278.1 KX443434 ASO98009.1 KJ645907 AIJ00764.1 KJ645905 AIJ00762.1 EF528485 ABP99019.1 BABH01022698 MH151918 QAX33066.1 KJ645908 AIJ00765.1 GU731530 ADE05580.1 KJ645909 AIJ00766.1 ODYU01004193 SOQ43795.1 ODYU01004192 SOQ43793.1
GQ241737 ADD70250.1 FJ387169 ACJ10074.1 EU309477 ABY40426.1 EF432657 ABO21683.1 EF547545 ABQ42339.1 EU273875 ABX64438.1 EF528486 ABP99020.1 BABH01022694 EF415296 ABN71367.1 AK289290 BAM73817.1 KU145393 ANC96676.1 GU731531 ADE05581.1 JTDY01009617 KOB63214.1 KX443435 ASO98010.1 JTDY01000942 KOB75351.1 NWSH01002736 PCG67605.1 KM016744 AID54896.1 DQ256407 ABB69054.1 KM016742 AID54894.1 KM016736 AID54888.1 KM016738 AID54890.1 JTDY01000272 KOB77971.1 KJ671575 AID55427.1 NWSH01003425 PCG66327.1 KM016741 AID54893.1 AY295774 AAP83689.1 GCVX01000058 JAI18172.1 KM016735 AID54887.1 JN375491 AEY75585.1 JN375489 AEY75583.1 JTDY01005369 KOB67074.1 KM016737 AID54889.1 GU937509 ADW23116.1 RSAL01000158 RVE45547.1 KR095600 AKS48888.1 DQ986461 JN176574 ABI84381.1 AET11931.1 NWSH01002369 PCG68453.1 PCG68452.1 KM016740 AID54892.1 KX443432 ASO98007.1 PCG67604.1 EU807990 ACF17813.2 KX443430 ASO98005.1 KJ671576 AID55428.1 ODYU01000082 SOQ34236.1 ODYU01008863 SOQ52829.1 PCG68451.1 JX310082 AFP20593.1 KP001126 AJN91171.1 GCVX01000081 JAI18149.1 RVE45548.1 FN356971 CAX94849.1 JX310078 AFP20589.1 BABH01022705 EF432656 ABO21682.1 RVE45549.1 KY212044 ARA91608.1 KY212043 ARA91607.1 KQ460655 KPJ12961.1 KY212045 ARA91609.1 JX310077 AFP20588.1 KM016739 AID54891.1 KF701137 AHW57307.1 MH236461 QBA57466.1 KJ645906 AIJ00763.1 ODYU01000083 SOQ34238.1 KQ459388 KPJ01135.1 KX443431 ASO98006.1 GQ915322 ACZ97416.2 KF853191 AHH92929.1 EU273877 ABX64440.1 RVE45546.1 EF535809 ABQ08711.1 KP001127 AJN91172.1 NWSH01000502 PCG75998.1 KX844829 ASQ44127.1 GCVX01000049 JAI18181.1 KZ150141 PZC72944.1 AK343206 BAM73892.1 FN544262 CBB07053.1 KM016743 AID54895.1 KX443433 ASO98008.1 KJ645904 AIJ00761.1 JX310083 AFP20594.1 ODYU01004424 SOQ44278.1 KX443434 ASO98009.1 KJ645907 AIJ00764.1 KJ645905 AIJ00762.1 EF528485 ABP99019.1 BABH01022698 MH151918 QAX33066.1 KJ645908 AIJ00765.1 GU731530 ADE05580.1 KJ645909 AIJ00766.1 ODYU01004193 SOQ43795.1 ODYU01004192 SOQ43793.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N766
L0N7C5
D6MLU9
B6VFR9
A9YXV8
A4GUB9
+ More
A5JM34 A9QW13 A5HG34 H9JUM4 A3RIC0 L0N6H5 A0A172MF86 D5L0M6 A0A0L7KJY7 A0A248QEX7 A0A0L7LJH7 A0A2A4J698 A0A068EWQ5 Q2XSW1 A0A068EU72 A0A068EVK6 A0A068F0X2 A0A0L7LRY0 A0A068EVD2 A0A2A4J2E3 A0A068EVL0 Q7YZS2 A0A0K8TUL3 A0A068ETU4 H2CZF1 H2CZE9 A0A0L7KVL1 A0A068EU65 F4YCW0 A0A3S2LFF7 A0A0K0YD30 Q069I9 A0A2A4JAM9 A0A2A4JAK3 A0A068ETU8 A0A248QHF4 A0A2A4J7V4 B3VT94 A0A248QEF8 A0A068F1Z7 A0A2H1V0E7 A0A2H1WID4 A0A2A4J918 J7FKN2 A0A0C5C1I6 A0A0K8TUI1 A0A3S2P9Y7 C1LZ54 J7FJF2 H9JUM8 A4GUB8 A0A437B5H6 A0A1V0D9E0 A0A1V0D9D9 A0A0N1IP03 A0A1V0D9D6 J7FKN1 A0A068EWQ0 X5D9B5 A0A411E5Q0 A0A0A7DH57 A0A2H1V048 A0A0N1ID06 A0A286QUG7 D2JLK6 W5VY13 A9QW15 A0A3S2LW02 A5HKM1 A0A0C5CGV6 A0A2A4JWC6 A0A222NX20 A0A0K8TV92 A0A2W1BGY9 L0N6M3 C7U1L4 A0A068F0X7 A0A248QG18 A0A0A7DH10 H9JUM5 J7FJF3 A0A2H1VU17 A0A248QEH8 A0A0A7DGX3 A0A0A7DH65 A5HG33 H9JUM6 A0A411AG09 A0A0A7DG86 D5L0M5 A0A0A7DH14 A0A2H1VSH4 A0A2H1VSG7
A5JM34 A9QW13 A5HG34 H9JUM4 A3RIC0 L0N6H5 A0A172MF86 D5L0M6 A0A0L7KJY7 A0A248QEX7 A0A0L7LJH7 A0A2A4J698 A0A068EWQ5 Q2XSW1 A0A068EU72 A0A068EVK6 A0A068F0X2 A0A0L7LRY0 A0A068EVD2 A0A2A4J2E3 A0A068EVL0 Q7YZS2 A0A0K8TUL3 A0A068ETU4 H2CZF1 H2CZE9 A0A0L7KVL1 A0A068EU65 F4YCW0 A0A3S2LFF7 A0A0K0YD30 Q069I9 A0A2A4JAM9 A0A2A4JAK3 A0A068ETU8 A0A248QHF4 A0A2A4J7V4 B3VT94 A0A248QEF8 A0A068F1Z7 A0A2H1V0E7 A0A2H1WID4 A0A2A4J918 J7FKN2 A0A0C5C1I6 A0A0K8TUI1 A0A3S2P9Y7 C1LZ54 J7FJF2 H9JUM8 A4GUB8 A0A437B5H6 A0A1V0D9E0 A0A1V0D9D9 A0A0N1IP03 A0A1V0D9D6 J7FKN1 A0A068EWQ0 X5D9B5 A0A411E5Q0 A0A0A7DH57 A0A2H1V048 A0A0N1ID06 A0A286QUG7 D2JLK6 W5VY13 A9QW15 A0A3S2LW02 A5HKM1 A0A0C5CGV6 A0A2A4JWC6 A0A222NX20 A0A0K8TV92 A0A2W1BGY9 L0N6M3 C7U1L4 A0A068F0X7 A0A248QG18 A0A0A7DH10 H9JUM5 J7FJF3 A0A2H1VU17 A0A248QEH8 A0A0A7DGX3 A0A0A7DH65 A5HG33 H9JUM6 A0A411AG09 A0A0A7DG86 D5L0M5 A0A0A7DH14 A0A2H1VSH4 A0A2H1VSG7
PDB
5VCG
E-value=5.27986e-52,
Score=518
Ontologies
GO
Topology
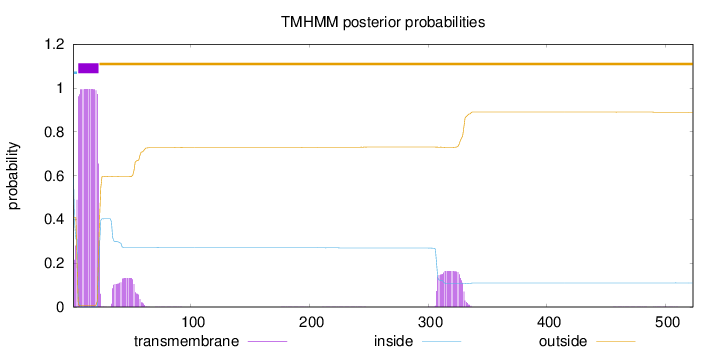
Length:
523
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.12513
Exp number, first 60 AAs:
21.43395
Total prob of N-in:
0.59126
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 22
outside
23 - 523
Population Genetic Test Statistics
Pi
277.292734
Theta
167.714848
Tajima's D
1.873104
CLR
0
CSRT
0.864756762161892
Interpretation
Uncertain
