Gene
KWMTBOMO09951 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013237
Annotation
cytochrome_P450_6ae2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.71 PlasmaMembrane Reliability : 1.401
Sequence
CDS
ATGTCAGTGAGCGCATTAGTATTTGCCGCCTTCGTTTTATTAGTGACGTATATTTACTATTGGTCGACCCGGAAATTCGATTATTGGAAGCGAAAAAATGTGCCATACGCTAAACCTGTGCCGTTCTTTGGGAATTACATGCGATATATCACTCTGCAGAGTTACATAGGAGATGTGACGCAGAAGCTGTGCCAGCAGTTCCCTGACAGACCGTACTTCGGATCGTTTTACGGCACGGAACCAGCCCTAGTCTTGCAGAATCCGGAGATCATCAAGCAAGTTTTTACTAAAGACTTCTATTACTTCAACAGCCGCGATAATAGTGATTACAATCATAAGGAAGTAGTCACACAGAACTTGTTCTTCGCAAATGGAGATAAATGGAAGGTAGTGCGTCAGAACCTAACACCGCTGTTCTCGTCGTTAAAGATGAAGAATATGTTTTACTTGGTGGAAAAATGCAACCATTCCCTTGAAGACATGCTGGACAAGGAAATCAAAGACTTGCAGATTATCGAGATCCGGTCTGCTATGATAAGGTACACGCTAGACTCTATCTGCTCTAGCGCCTTCGGTATTGAAAGTAATACCTTGAGAGAGGGCGGAGAAAATAGTCCTTTTGCAAACATGGGAAGCATCGTATTTAGTTCTTCAATAACTCGAGGACTGAAATGGATCAGTCGTTCGATGTGGCCGGGCATATTCTACAAGCTTGGTCTCCAGTGCTTTCCAGCGGAGATTGACGGTTTCTTTGAGAGAGTCCTGATTGAGGTGTTCGAGAACCGTGGCTACAAGCCGACGAACCGAAACGATTTCGTGGACTTAATTTTGAGTCTGAAACAAAACGATTACCTGACCGGAGATGGCTTGGTTCCCAAAAACGTGGACGCAAAGAAAGTGACGGTCAAGGTTGACGACGCCTTATTGATAGCGCAGTGCGTAGTCTTCTTTGGCGCAGGCTTCGAAACGTCAGCTAGCACTCTCTCGGCAGCCCTATACGAACTGGCCAAGAACCCAGAAGCCCAAAGGCGCGCTCAAGAGGAAGTTGACGAACTTTTGCTAAAACACAACAACAAGTTAAATTATGATTGTTTAGCGGAATTGCCGTACTTGGAGGCATGTATGAACGAAGCGATGCGTCTGTACCCAGTACTTCCTAATATCACCCGCGAAGCCGTGACCGACTATACGTTTCCTGATGGTCTGAGGATAGATAAGGGTATGCGTGTGCATGTCCCGGTGTATGCTATCCACCGTAACCCCGACAACTTTCCTGACCCGGAAGAGTTCCGTCCTGAGAGACATTTAGGAGACGCCAAAAACGACATTAAGCAGTTCACGTACTTCCCGTTCGGAGAAGGGCCCAGGATCTGTATTGGTATGCGCTTCGGGAAAATGCAGACCATCGCTGGGATGGTTACCTGTTTGAAAAAGTACAACTTCGAACTGGCGGATGGTATGTCGAAAACGGTGCCCTTCAGATCCACAACAGTGCTAACACAGCCAAGTACCGGATTATTCCTGAAGGCCACACCACGAGACGGCTGGAAGCAACGTCCGAACAACAACATTCGGCACTTCGATAAAGCGGCACGACACGAGAACCCTCTCATCGTGGCCGCCGGTAACTACATTCCCGATCCTGCGGACAGAATGGAAAGCAGTCGACGTCGCCCAAAACACGTCATCTCGGATCCTCCCGATCCACTAACGGTGCTTTTAGGGTATATCCTACTAAAGGAAAGTATATCCAAAGTGGTGCATAATTTGTGCAAATTATTTCCAAATGACCCCTACATTGGTGCGTTCTTTGGCACAGAGCCGACTTTAATAGTTAAAGACCCGGAATTCATTAAACTGGTATTAACTAAAGATTTCTACCATTTCAACGGCAGAGAAGGCTCCAAATACACCCACAATGAAGTTGTAACACAGAACATATTTTTCACGTACGGTGATCGGTGGAAGGTCATCCGGCAAAACCTGACGCCGCTTTTCTCTTCTTTAAAAATGAGAAACATGTTTCATATTATCGAGAAATGTTCTGGTATTTTCGAGAATTTGCTGGATGAAGAATCTTTAGCACCCGAAGTTGAAATGAAGTCTTTAATGTCGCGGTTCACAATGGACTGCATAGGCGGTTGTGCGTTCGGAGTAGACACTAAAGCGATGCAGGAACCCAAGGATAACATCTTCACAACTATGGGATACCTTTTCTTCGAGAGCACCACCTACCGAGGGATCAAGAACGTTCTCAGAGCCATTTGGCCAGGGATATTCTATGGACTAGGTCTCAAAGTATTTCCAACGGATTTAAATGAGTTTTTTAGTAAACTTCTAGTAGGGATTTTCGAAGCACGGGACTACAAGCCTTCCTCACGGAACGATTTCGTTGACCTGTTGTTAAATTTGAAGAAGAACAGACATATCGTCGGCGACCGTTTGCAAAAGACTACAACAGGAGACGAAGGAGCAGACAGTAAATTTGAACTGGAAGTGGATGATGGGCTTTTAGTGGGGCAATGCCTAGCCTTCTTTAGCGCAGGTTTTGAAACTACATCCACTATTTCAAACTTTACTTTGTACGAGCTGGCAAAGAATCCAGATGTTCAGAAAAGAGCACAGAAAGAAGTAGACGAATATATTAAAAAGCACAACAATAAACTAGATTATGATTGCGTGAAGGAGCTCCCGTTTGTGGAGGCGTGTATAGATGAAGCGCTACGCTTGTATCCTGTTCTTGGAGTATTGACAAGAGAAGTAATGGAACAATACACATTCCCGACCGGCCTGACTCTCGATAAAGGAGATCGCGTCCATATTCCCGTGTATCATCTGCAACGCGACCCGGAATACTTTCCGGAGCCAGAGCTCTTCAAGCCGGAGAGGTTTTATGGGGAAGAAAAGAAGAATATCAGACCCTTTACGTATTTGCCATTTGGGGACGGACCACGCATTTGTATTGGTATGAGGTTCGCGAAGATGCAGATCCTGGCCGGTCTGGTGACAATACTGAAAAAGTACACAGTGCAACTGGCCGACGGTATGCCGGAGACGATCGATATCGAACCCAAAGCCATTGTTACGCAGCCAGCCATTAGCATGCGACTAAAATTCGTTCCGAGAAATGATTTGCAGAAAAGAATTTTCGCATAG
Protein
MSVSALVFAAFVLLVTYIYYWSTRKFDYWKRKNVPYAKPVPFFGNYMRYITLQSYIGDVTQKLCQQFPDRPYFGSFYGTEPALVLQNPEIIKQVFTKDFYYFNSRDNSDYNHKEVVTQNLFFANGDKWKVVRQNLTPLFSSLKMKNMFYLVEKCNHSLEDMLDKEIKDLQIIEIRSAMIRYTLDSICSSAFGIESNTLREGGENSPFANMGSIVFSSSITRGLKWISRSMWPGIFYKLGLQCFPAEIDGFFERVLIEVFENRGYKPTNRNDFVDLILSLKQNDYLTGDGLVPKNVDAKKVTVKVDDALLIAQCVVFFGAGFETSASTLSAALYELAKNPEAQRRAQEEVDELLLKHNNKLNYDCLAELPYLEACMNEAMRLYPVLPNITREAVTDYTFPDGLRIDKGMRVHVPVYAIHRNPDNFPDPEEFRPERHLGDAKNDIKQFTYFPFGEGPRICIGMRFGKMQTIAGMVTCLKKYNFELADGMSKTVPFRSTTVLTQPSTGLFLKATPRDGWKQRPNNNIRHFDKAARHENPLIVAAGNYIPDPADRMESSRRRPKHVISDPPDPLTVLLGYILLKESISKVVHNLCKLFPNDPYIGAFFGTEPTLIVKDPEFIKLVLTKDFYHFNGREGSKYTHNEVVTQNIFFTYGDRWKVIRQNLTPLFSSLKMRNMFHIIEKCSGIFENLLDEESLAPEVEMKSLMSRFTMDCIGGCAFGVDTKAMQEPKDNIFTTMGYLFFESTTYRGIKNVLRAIWPGIFYGLGLKVFPTDLNEFFSKLLVGIFEARDYKPSSRNDFVDLLLNLKKNRHIVGDRLQKTTTGDEGADSKFELEVDDGLLVGQCLAFFSAGFETTSTISNFTLYELAKNPDVQKRAQKEVDEYIKKHNNKLDYDCVKELPFVEACIDEALRLYPVLGVLTREVMEQYTFPTGLTLDKGDRVHIPVYHLQRDPEYFPEPELFKPERFYGEEKKNIRPFTYLPFGDGPRICIGMRFAKMQILAGLVTILKKYTVQLADGMPETIDIEPKAIVTQPAISMRLKFVPRNDLQKRIFA
Summary
Uniprot
EMBL
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
Ontologies
GO
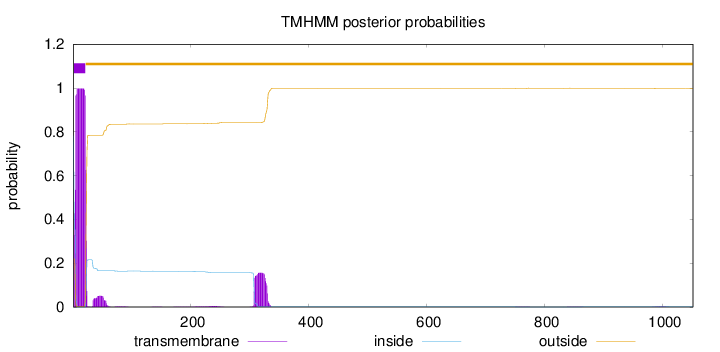
Topology
Length:
1051
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.8042999999999
Exp number, first 60 AAs:
20.15986
Total prob of N-in:
0.78134
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 1051
Population Genetic Test Statistics
Pi
197.45438
Theta
147.875942
Tajima's D
1.266131
CLR
0.055925
CSRT
0.738313084345783
Interpretation
Uncertain
