Gene
KWMTBOMO09941
Pre Gene Modal
BGIBMGA013242
Annotation
cis?_cis-muconate_transporter_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.956
Sequence
CDS
ATGTTCCGCAGCGGTCTACTGGGTAAGCTGGGCGACTTCTACAAATGGTACGTCCTCGGTCTTGTCTCCGTCGGCTACATATTGGGAGAACTTGGACATTATCTCATAGGTACGACATCAAAAGTCACAGCGGAAGACCTTCATTACGGCGACAAGTCTTGCATGCTGAATGCCACACATCTGTCGCTTAGCCAACTTCCAGTTGTCTGTGAGAAGGTTAATTCCTCTGAAATATGCGAGTCCTTAACTCTGAACGGTACACGGTACTGCGAATGGGGCTACAATGGCCTCGGCATCGACTATCAAGTCCTGGCTGGACCTGCCTTCATGGCGGTCTTCACCGTTGTCGGAGTTATACTTGGAGTGGTCGCGGACAAATATAATAGGGCACGCATTCTCTCGATTTGCACTCTGGTTTTCGTTGTCGCGATTCTCCTCATGGGCACGGTGACAGAATACTGGCATCTCGTAGTACTCCGTATGGTCATGGCGGCAGGGGAATCAGGCTGCAATCCCTTGGCGACAGGAATCCTGACAGATCTGTTCGCGGAGCATCAGAGAGCCTTGGTTTTGTCCATCTTCAACTGGGGTATTTATGGGGGATACGGTATAGCTTTCCCGGTTGGAAGGTACATCCCTGACTTGAATATTTGGGGTCTGAACTGGCGCTTGTGCTACTACGGAGCTGGAGTCGTGGGTCTGGTGATCGCTGCCCTGACGTTCCTCACGCTTAAGGAGCCTGAACGTACCACCATCGGTGAAGAAGGTAATACGAAAAGCAACGACGCAACTCTGGAGTCTGGTAAAAAGGTGCCGCAGGTCACAATCTGGCACGTCATCTCCCAGCCCAGGATACTGCTCCTCTGCTTAGCTGCTTCGATCAGACATTGTGGTGGTATGTGCTTCGCCTACAATGCCGATCTGTACTACCGTGACTACTTCCCCGACGTCGATCTCGGCTGGTGGCTGTTCGCTGTCACGGTCGGCATTGGATCCGTTGGTGTTGTCGCCGGCGGAATTATATCGGACAAATACGTAGCGAAAATGGGGATTAGGTCCCGAGTGCTTGTGCTCGCGTTGTCCCAGCTGATAGCTACCTTACCTTCGTTCGGGTCCGTCGTCTTCGGCCCGCTTTGGGCTATGATCACACTGGCCTTCGCATATTTCTTTGCCGAGATGTGGTTCGGCATTGTGTTCGCGGTATTGGTGGAGATCGTGCCCTTGTCCGTGCGTTCCACAACCGTCGGCGTCTTCCTCTTTGTTATGAACAACATCGGAGGAAATCTGCCAATTCTGGTCGACCCCGTTTCCAAGGCCATCGGTTATAGAGAAGCCATCATGATTTTCTATGCCGGATTTTATGGCATCAGCAGCATTCTGTTCTTCTTGACAATGTTCCTGATGGACGGACCGGTTGAGAAACAGGAAGAAACCACGACACCCACAGGATTGGACAACAGAGCTTACAGCCACGACGAGCTCAGGCAAACGAGAGCATTACCTAACTCACCTAACAGTAGATTATAA
Protein
MFRSGLLGKLGDFYKWYVLGLVSVGYILGELGHYLIGTTSKVTAEDLHYGDKSCMLNATHLSLSQLPVVCEKVNSSEICESLTLNGTRYCEWGYNGLGIDYQVLAGPAFMAVFTVVGVILGVVADKYNRARILSICTLVFVVAILLMGTVTEYWHLVVLRMVMAAGESGCNPLATGILTDLFAEHQRALVLSIFNWGIYGGYGIAFPVGRYIPDLNIWGLNWRLCYYGAGVVGLVIAALTFLTLKEPERTTIGEEGNTKSNDATLESGKKVPQVTIWHVISQPRILLLCLAASIRHCGGMCFAYNADLYYRDYFPDVDLGWWLFAVTVGIGSVGVVAGGIISDKYVAKMGIRSRVLVLALSQLIATLPSFGSVVFGPLWAMITLAFAYFFAEMWFGIVFAVLVEIVPLSVRSTTVGVFLFVMNNIGGNLPILVDPVSKAIGYREAIMIFYAGFYGISSILFFLTMFLMDGPVEKQEETTTPTGLDNRAYSHDELRQTRALPNSPNSRL
Summary
Similarity
Belongs to the ephrin family.
Uniprot
G8GB89
H9JUM9
A0A437B4W6
G8GB90
A0A212FJ45
A0A0N1PFV8
+ More
A0A0N1PFR2 A0A2A4JNH8 A0A2H1VQ72 E9IEL3 E2B4H1 A0A088A8H3 A0A2A3EGH7 A0A310SC39 A0A154P7V8 A0A0L7REA7 A0A151WSR3 A0A195FQS7 A0A2J7R842 A0A195CCW7 W8JTP3 A0A158NE87 F4WJE1 A0A232F0Q9 A0A026WYZ5 K7J947 B4N1G3 A0A1Y1LCF7 A0A0C9RGX1 A0A1L8DEI2 B3MS96 B4L6C5 A0A067QZG5 A0A3B0JX55 A0A1W4VEV1 B4MAE3 Q29IM2 B4PZ75 A0A0R3NYQ2 A0A0R1E9U0 B3NXV4 B4JK24 B4I062 Q9W3W7 C3KGN5 A0A0L0CKV0 A0A182PSG0 A0A182S8Y2 A0A182QUB5 A0A182TZM6 A0A1I8PAW5 A0A182X9N3 A0A1B0GM24 A0A182HKL7 A0A182VFC5 A0A182L8V9 Q7PUR7 A0A195BXA1 A0A0J7KVW9 A0A0A1WRQ6 A0A182K9T6 W8C218 A0A182VR44 A0A182MMS7 W8BFQ3 A0A182NSC5 A0A0M4F7V6 A0A182YRJ3 A0A182RXI6 A0A182F871 W5JAN9 A0A084VS63 A0A182IR50 A0A023EW47 A0A1Q3EYP5 A0A0K8VMC9 Q16PZ8 B0X7V1 A0A1I8MZT5 A0A1A9XSU5 A0A1B0BTW8 A0A1B0GFF1 A0A1A9UFF7 A0A1A9ZLE7 A0A1A9WA07 A0A3B0KRV7 A0A1W4WI15 A0A1J1INU2 A0A2P8XYV1 E0V9B1 E2AQ68 B4JJZ5 A0A1B6MGD8 J9JNI4 A0A2H8TP15 A0A146LGV6 A0A1L8DE20 D2A5V2 A0A1W4WTV4 N6T6C0
A0A0N1PFR2 A0A2A4JNH8 A0A2H1VQ72 E9IEL3 E2B4H1 A0A088A8H3 A0A2A3EGH7 A0A310SC39 A0A154P7V8 A0A0L7REA7 A0A151WSR3 A0A195FQS7 A0A2J7R842 A0A195CCW7 W8JTP3 A0A158NE87 F4WJE1 A0A232F0Q9 A0A026WYZ5 K7J947 B4N1G3 A0A1Y1LCF7 A0A0C9RGX1 A0A1L8DEI2 B3MS96 B4L6C5 A0A067QZG5 A0A3B0JX55 A0A1W4VEV1 B4MAE3 Q29IM2 B4PZ75 A0A0R3NYQ2 A0A0R1E9U0 B3NXV4 B4JK24 B4I062 Q9W3W7 C3KGN5 A0A0L0CKV0 A0A182PSG0 A0A182S8Y2 A0A182QUB5 A0A182TZM6 A0A1I8PAW5 A0A182X9N3 A0A1B0GM24 A0A182HKL7 A0A182VFC5 A0A182L8V9 Q7PUR7 A0A195BXA1 A0A0J7KVW9 A0A0A1WRQ6 A0A182K9T6 W8C218 A0A182VR44 A0A182MMS7 W8BFQ3 A0A182NSC5 A0A0M4F7V6 A0A182YRJ3 A0A182RXI6 A0A182F871 W5JAN9 A0A084VS63 A0A182IR50 A0A023EW47 A0A1Q3EYP5 A0A0K8VMC9 Q16PZ8 B0X7V1 A0A1I8MZT5 A0A1A9XSU5 A0A1B0BTW8 A0A1B0GFF1 A0A1A9UFF7 A0A1A9ZLE7 A0A1A9WA07 A0A3B0KRV7 A0A1W4WI15 A0A1J1INU2 A0A2P8XYV1 E0V9B1 E2AQ68 B4JJZ5 A0A1B6MGD8 J9JNI4 A0A2H8TP15 A0A146LGV6 A0A1L8DE20 D2A5V2 A0A1W4WTV4 N6T6C0
Pubmed
22885739
19121390
22118469
26354079
21282665
20798317
+ More
24681434 21347285 21719571 28648823 24508170 30249741 20075255 17994087 28004739 24845553 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 20966253 12364791 14747013 17210077 25830018 24495485 25244985 20920257 23761445 24438588 24945155 26483478 17510324 25315136 29403074 20566863 26823975 18362917 19820115 23537049
24681434 21347285 21719571 28648823 24508170 30249741 20075255 17994087 28004739 24845553 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 20966253 12364791 14747013 17210077 25830018 24495485 25244985 20920257 23761445 24438588 24945155 26483478 17510324 25315136 29403074 20566863 26823975 18362917 19820115 23537049
EMBL
JN118491
AB610783
AER70323.1
BAM34533.1
BABH01022711
BABH01022712
+ More
RSAL01000158 RVE45550.1 JN118492 AB610784 AER70324.1 BAM34534.1 AGBW02008320 OWR53730.1 KQ460655 KPJ12962.1 KQ459388 KPJ01133.1 NWSH01001037 PCG72993.1 ODYU01003791 SOQ42993.1 GL762647 EFZ20984.1 GL445560 EFN89407.1 KZ288254 PBC30883.1 KQ772169 OAD52486.1 KQ434822 KZC07210.1 KQ414609 KOC69307.1 KQ982769 KYQ50863.1 KQ981335 KYN42652.1 NEVH01006723 PNF36998.1 KQ978023 KYM97928.1 KF951599 AHK59785.1 ADTU01013173 ADTU01013174 GL888182 EGI65719.1 NNAY01001379 OXU24172.1 KK107078 QOIP01000011 EZA60364.1 RLU17229.1 CH963925 EDW78202.1 GEZM01060073 JAV71303.1 GBYB01007580 JAG77347.1 GFDF01009215 JAV04869.1 CH902622 EDV34651.1 CH933812 EDW05921.1 KK853156 KDR10504.1 OUUW01000011 SPP86657.1 CH940655 EDW66202.1 CH379063 EAL32631.1 CM000162 EDX01042.2 KRT06190.1 KRK05918.1 KRK05919.1 CH954180 EDV47405.1 CH916370 EDV99926.1 CH480819 EDW52893.1 AE014298 BT016060 BT082129 AAF46196.1 AAV36945.1 ACQ57836.1 BT082100 ACQ41875.1 JRES01000262 KNC32890.1 AXCN02000207 AJVK01020232 APCN01000846 AAAB01008987 EAA01128.5 KQ976401 KYM92581.1 LBMM01002757 KMQ94409.1 GBXI01012765 GBXI01002588 GBXI01000458 JAD01527.1 JAD11704.1 JAD13834.1 GAMC01010781 JAB95774.1 AXCM01007595 GAMC01010782 JAB95773.1 CP012528 ALC48350.1 ADMH02002026 ETN59900.1 ATLV01015825 KE525036 KFB40807.1 JXUM01043170 GAPW01001014 KQ561353 JAC12584.1 KXJ78859.1 GFDL01014636 JAV20409.1 GDHF01012609 JAI39705.1 CH477768 EAT36431.1 DS232464 EDS42170.1 JXJN01020406 CCAG010013954 SPP86658.1 CVRI01000057 CRL01903.1 PYGN01001149 PSN37166.1 DS234991 EEB09967.1 GL441701 EFN64402.1 EDV99897.1 GEBQ01018183 GEBQ01004960 JAT21794.1 JAT35017.1 ABLF02033967 GFXV01004118 MBW15923.1 GDHC01012899 JAQ05730.1 GFDF01009459 JAV04625.1 KQ971345 EFA05422.1 APGK01050470 APGK01050471 KB741174 ENN73218.1
RSAL01000158 RVE45550.1 JN118492 AB610784 AER70324.1 BAM34534.1 AGBW02008320 OWR53730.1 KQ460655 KPJ12962.1 KQ459388 KPJ01133.1 NWSH01001037 PCG72993.1 ODYU01003791 SOQ42993.1 GL762647 EFZ20984.1 GL445560 EFN89407.1 KZ288254 PBC30883.1 KQ772169 OAD52486.1 KQ434822 KZC07210.1 KQ414609 KOC69307.1 KQ982769 KYQ50863.1 KQ981335 KYN42652.1 NEVH01006723 PNF36998.1 KQ978023 KYM97928.1 KF951599 AHK59785.1 ADTU01013173 ADTU01013174 GL888182 EGI65719.1 NNAY01001379 OXU24172.1 KK107078 QOIP01000011 EZA60364.1 RLU17229.1 CH963925 EDW78202.1 GEZM01060073 JAV71303.1 GBYB01007580 JAG77347.1 GFDF01009215 JAV04869.1 CH902622 EDV34651.1 CH933812 EDW05921.1 KK853156 KDR10504.1 OUUW01000011 SPP86657.1 CH940655 EDW66202.1 CH379063 EAL32631.1 CM000162 EDX01042.2 KRT06190.1 KRK05918.1 KRK05919.1 CH954180 EDV47405.1 CH916370 EDV99926.1 CH480819 EDW52893.1 AE014298 BT016060 BT082129 AAF46196.1 AAV36945.1 ACQ57836.1 BT082100 ACQ41875.1 JRES01000262 KNC32890.1 AXCN02000207 AJVK01020232 APCN01000846 AAAB01008987 EAA01128.5 KQ976401 KYM92581.1 LBMM01002757 KMQ94409.1 GBXI01012765 GBXI01002588 GBXI01000458 JAD01527.1 JAD11704.1 JAD13834.1 GAMC01010781 JAB95774.1 AXCM01007595 GAMC01010782 JAB95773.1 CP012528 ALC48350.1 ADMH02002026 ETN59900.1 ATLV01015825 KE525036 KFB40807.1 JXUM01043170 GAPW01001014 KQ561353 JAC12584.1 KXJ78859.1 GFDL01014636 JAV20409.1 GDHF01012609 JAI39705.1 CH477768 EAT36431.1 DS232464 EDS42170.1 JXJN01020406 CCAG010013954 SPP86658.1 CVRI01000057 CRL01903.1 PYGN01001149 PSN37166.1 DS234991 EEB09967.1 GL441701 EFN64402.1 EDV99897.1 GEBQ01018183 GEBQ01004960 JAT21794.1 JAT35017.1 ABLF02033967 GFXV01004118 MBW15923.1 GDHC01012899 JAQ05730.1 GFDF01009459 JAV04625.1 KQ971345 EFA05422.1 APGK01050470 APGK01050471 KB741174 ENN73218.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000008237 UP000005203 UP000242457 UP000076502 UP000053825 UP000075809 UP000078541 UP000235965 UP000078542 UP000005205 UP000007755 UP000215335 UP000053097 UP000279307 UP000002358 UP000007798 UP000007801 UP000009192 UP000027135 UP000268350 UP000192221 UP000008792 UP000001819 UP000002282 UP000008711 UP000001070 UP000001292 UP000000803 UP000037069 UP000075885 UP000075901 UP000075886 UP000075902 UP000095300 UP000076407 UP000092462 UP000075840 UP000075903 UP000075882 UP000007062 UP000078540 UP000036403 UP000075881 UP000075920 UP000075883 UP000075884 UP000092553 UP000076408 UP000075900 UP000069272 UP000000673 UP000030765 UP000075880 UP000069940 UP000249989 UP000008820 UP000002320 UP000095301 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000091820 UP000192223 UP000183832 UP000245037 UP000009046 UP000000311 UP000007819 UP000007266 UP000019118
UP000008237 UP000005203 UP000242457 UP000076502 UP000053825 UP000075809 UP000078541 UP000235965 UP000078542 UP000005205 UP000007755 UP000215335 UP000053097 UP000279307 UP000002358 UP000007798 UP000007801 UP000009192 UP000027135 UP000268350 UP000192221 UP000008792 UP000001819 UP000002282 UP000008711 UP000001070 UP000001292 UP000000803 UP000037069 UP000075885 UP000075901 UP000075886 UP000075902 UP000095300 UP000076407 UP000092462 UP000075840 UP000075903 UP000075882 UP000007062 UP000078540 UP000036403 UP000075881 UP000075920 UP000075883 UP000075884 UP000092553 UP000076408 UP000075900 UP000069272 UP000000673 UP000030765 UP000075880 UP000069940 UP000249989 UP000008820 UP000002320 UP000095301 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000091820 UP000192223 UP000183832 UP000245037 UP000009046 UP000000311 UP000007819 UP000007266 UP000019118
Interpro
Gene 3D
CDD
ProteinModelPortal
G8GB89
H9JUM9
A0A437B4W6
G8GB90
A0A212FJ45
A0A0N1PFV8
+ More
A0A0N1PFR2 A0A2A4JNH8 A0A2H1VQ72 E9IEL3 E2B4H1 A0A088A8H3 A0A2A3EGH7 A0A310SC39 A0A154P7V8 A0A0L7REA7 A0A151WSR3 A0A195FQS7 A0A2J7R842 A0A195CCW7 W8JTP3 A0A158NE87 F4WJE1 A0A232F0Q9 A0A026WYZ5 K7J947 B4N1G3 A0A1Y1LCF7 A0A0C9RGX1 A0A1L8DEI2 B3MS96 B4L6C5 A0A067QZG5 A0A3B0JX55 A0A1W4VEV1 B4MAE3 Q29IM2 B4PZ75 A0A0R3NYQ2 A0A0R1E9U0 B3NXV4 B4JK24 B4I062 Q9W3W7 C3KGN5 A0A0L0CKV0 A0A182PSG0 A0A182S8Y2 A0A182QUB5 A0A182TZM6 A0A1I8PAW5 A0A182X9N3 A0A1B0GM24 A0A182HKL7 A0A182VFC5 A0A182L8V9 Q7PUR7 A0A195BXA1 A0A0J7KVW9 A0A0A1WRQ6 A0A182K9T6 W8C218 A0A182VR44 A0A182MMS7 W8BFQ3 A0A182NSC5 A0A0M4F7V6 A0A182YRJ3 A0A182RXI6 A0A182F871 W5JAN9 A0A084VS63 A0A182IR50 A0A023EW47 A0A1Q3EYP5 A0A0K8VMC9 Q16PZ8 B0X7V1 A0A1I8MZT5 A0A1A9XSU5 A0A1B0BTW8 A0A1B0GFF1 A0A1A9UFF7 A0A1A9ZLE7 A0A1A9WA07 A0A3B0KRV7 A0A1W4WI15 A0A1J1INU2 A0A2P8XYV1 E0V9B1 E2AQ68 B4JJZ5 A0A1B6MGD8 J9JNI4 A0A2H8TP15 A0A146LGV6 A0A1L8DE20 D2A5V2 A0A1W4WTV4 N6T6C0
A0A0N1PFR2 A0A2A4JNH8 A0A2H1VQ72 E9IEL3 E2B4H1 A0A088A8H3 A0A2A3EGH7 A0A310SC39 A0A154P7V8 A0A0L7REA7 A0A151WSR3 A0A195FQS7 A0A2J7R842 A0A195CCW7 W8JTP3 A0A158NE87 F4WJE1 A0A232F0Q9 A0A026WYZ5 K7J947 B4N1G3 A0A1Y1LCF7 A0A0C9RGX1 A0A1L8DEI2 B3MS96 B4L6C5 A0A067QZG5 A0A3B0JX55 A0A1W4VEV1 B4MAE3 Q29IM2 B4PZ75 A0A0R3NYQ2 A0A0R1E9U0 B3NXV4 B4JK24 B4I062 Q9W3W7 C3KGN5 A0A0L0CKV0 A0A182PSG0 A0A182S8Y2 A0A182QUB5 A0A182TZM6 A0A1I8PAW5 A0A182X9N3 A0A1B0GM24 A0A182HKL7 A0A182VFC5 A0A182L8V9 Q7PUR7 A0A195BXA1 A0A0J7KVW9 A0A0A1WRQ6 A0A182K9T6 W8C218 A0A182VR44 A0A182MMS7 W8BFQ3 A0A182NSC5 A0A0M4F7V6 A0A182YRJ3 A0A182RXI6 A0A182F871 W5JAN9 A0A084VS63 A0A182IR50 A0A023EW47 A0A1Q3EYP5 A0A0K8VMC9 Q16PZ8 B0X7V1 A0A1I8MZT5 A0A1A9XSU5 A0A1B0BTW8 A0A1B0GFF1 A0A1A9UFF7 A0A1A9ZLE7 A0A1A9WA07 A0A3B0KRV7 A0A1W4WI15 A0A1J1INU2 A0A2P8XYV1 E0V9B1 E2AQ68 B4JJZ5 A0A1B6MGD8 J9JNI4 A0A2H8TP15 A0A146LGV6 A0A1L8DE20 D2A5V2 A0A1W4WTV4 N6T6C0
PDB
4LDS
E-value=0.00307567,
Score=97
Ontologies
GO
Topology
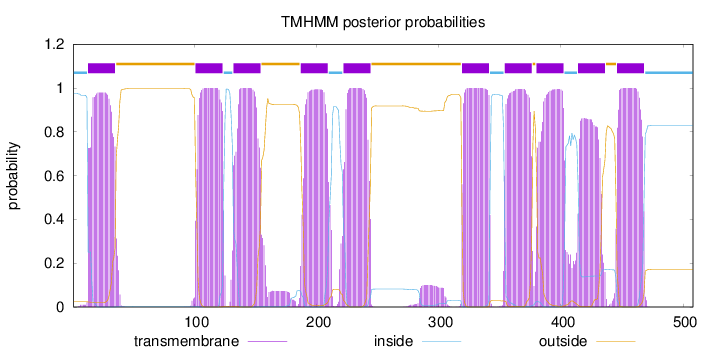
Length:
508
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
221.34725
Exp number, first 60 AAs:
21.6244
Total prob of N-in:
0.97628
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 100
TMhelix
101 - 123
inside
124 - 131
TMhelix
132 - 154
outside
155 - 186
TMhelix
187 - 209
inside
210 - 221
TMhelix
222 - 244
outside
245 - 318
TMhelix
319 - 341
inside
342 - 353
TMhelix
354 - 376
outside
377 - 379
TMhelix
380 - 402
inside
403 - 413
TMhelix
414 - 436
outside
437 - 445
TMhelix
446 - 468
inside
469 - 508
Population Genetic Test Statistics
Pi
279.584983
Theta
191.42634
Tajima's D
1.699461
CLR
0.043171
CSRT
0.828458577071146
Interpretation
Uncertain
