Gene
KWMTBOMO09940
Pre Gene Modal
BGIBMGA013182
Annotation
uncharacterized_protein_LOC778477_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.868
Sequence
CDS
ATGTCAGATTCATCTGAAGATGAAGACCTATCGCGTTTCAAAGACGTCGTCGACAACTCTTTCGTTAAATCTCTATCCCAAACTCGTGTTACAGGATCTAAAACAATTCAAGGCATTGAAGAAAAGCCCGTCTCCCAACGTTACTTAGAAATATCAAGCCACTACAATGATGTGAAGGTTCCAGAAGAAATGCAAAAAAGAATTGGTGCTAAATTATCAGCTGTCATACAGAAAAATACAAGATTTGTAGATGTGGACCCTGTTCAACCAAAAAAACGCAAAATAAAGGGAGGTGTAAAATTGTTCAAGAATTCAAATGGATTTTTATCTTGTGATGACATTAAGGACATATCCACTGAGAAACATAATTGTGAAGCGAAACTGTTAAAGTATAGTAAAGTTGGTCCTGACTCAGATGACATAGAAATATCTAAATCTGCTGTGGAGTAG
Protein
MSDSSEDEDLSRFKDVVDNSFVKSLSQTRVTGSKTIQGIEEKPVSQRYLEISSHYNDVKVPEEMQKRIGAKLSAVIQKNTRFVDVDPVQPKKRKIKGGVKLFKNSNGFLSCDDIKDISTEKHNCEAKLLKYSKVGPDSDDIEISKSAVE
Summary
Similarity
Belongs to the class-III pyridoxal-phosphate-dependent aminotransferase family.
Uniprot
EMBL
DQ813504
ABG67690.1
ODYU01011504
SOQ57275.1
NWSH01001037
PCG72992.1
+ More
GDQN01005561 JAT85493.1 RSAL01000158 RVE45551.1 KQ460655 KPJ12963.1 AGBW02008320 OWR53731.1 KQ459388 KPJ01131.1 GAIX01003687 JAA88873.1 JXUM01041642 KQ561292 KXJ79093.1 GFDL01011568 JAV23477.1 CH477729 EAT36792.1 EAT36794.1 APGK01019088 KB740092 ENN81434.1 DS232386 EDS40957.1 BDDW01000010 GAT88329.1
GDQN01005561 JAT85493.1 RSAL01000158 RVE45551.1 KQ460655 KPJ12963.1 AGBW02008320 OWR53731.1 KQ459388 KPJ01131.1 GAIX01003687 JAA88873.1 JXUM01041642 KQ561292 KXJ79093.1 GFDL01011568 JAV23477.1 CH477729 EAT36792.1 EAT36794.1 APGK01019088 KB740092 ENN81434.1 DS232386 EDS40957.1 BDDW01000010 GAT88329.1
Proteomes
PRIDE
Pfam
PF00202 Aminotran_3
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
Ontologies
PANTHER
Topology
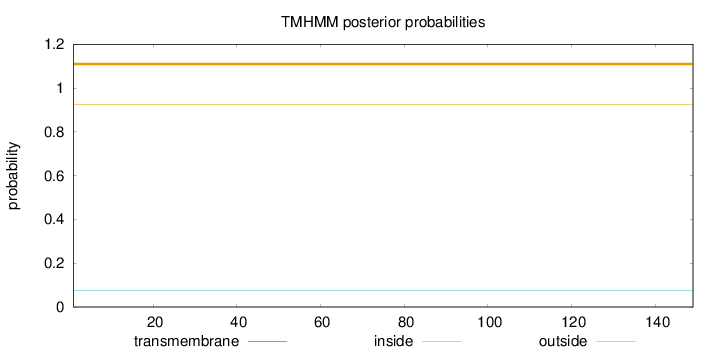
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07412
outside
1 - 149
Population Genetic Test Statistics
Pi
298.04026
Theta
181.478224
Tajima's D
2.048253
CLR
0
CSRT
0.893955302234888
Interpretation
Uncertain
