Gene
KWMTBOMO09939
Pre Gene Modal
BGIBMGA013243
Annotation
PREDICTED:_mitochondrial_import_inner_membrane_translocase_subunit_Tim22_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 2.263
Sequence
CDS
ATGGGTGAATACCGGAGACCACCTCCAGACCAAAATTTAGTGAACTTTACCCAAAATAGTGATTATGATTCGCTTGCAAAATATTTGATAGGCAACAACTACAGATTTCGTGAAAATATCATTATACCAAGAATTCTTGGACCTGTTATAATTAAAACCAATGAGGAGAAGATGATCGAGGCAACGGTTGAGAGCTGTCCCTTTAAATCATTGACTAGTTGTATAATTGGATATGGTCTCGGAGCAGCAGTGGGTCTATTCACATCTTCCCTGATGCCTAACGTAACAGATACTACTGCCCAACAAAATCAAACGGCTAGAGAGATTTTACGGGAAATGAAAAACTCCATGTTGAGTTATGGAAAAAACTTTGCGATACTTGGTGCTGTGTTTTCTGGGGTCGAGTGTTGTATCGAAACAGCAAGGGGGAAGTCCGACTGGAAGAACGGTACTTATGCTGGAGGAATTACAGGAGGATTAATTGGATTAAGAGGTGGATTAAAAGCTGGCATGTTTGGAGCGATGGGATTTGCAGCCTTTTCCACGATCATTGACTACTATATGCATCAACGGGCTTAA
Protein
MGEYRRPPPDQNLVNFTQNSDYDSLAKYLIGNNYRFRENIIIPRILGPVIIKTNEEKMIEATVESCPFKSLTSCIIGYGLGAAVGLFTSSLMPNVTDTTAQQNQTAREILREMKNSMLSYGKNFAILGAVFSGVECCIETARGKSDWKNGTYAGGITGGLIGLRGGLKAGMFGAMGFAAFSTIIDYYMHQRA
Summary
Uniprot
H9JUN0
A0A1E1WHR2
A0A0N0PC46
A0A212FJ28
A0A0N0P9V2
A0A2A4JLY3
+ More
A0A2H1WP47 A0A0L7KG57 A0A3S2NA46 A0A1A9YDQ5 B0X2Y3 A0A023EJD9 Q17Q11 A0A1A9UTG7 A0A1B0G195 A0A1B0B321 A0A2M4ANM6 W5JHI4 A0A1A9ZSR9 T1E7Y6 A0A2M4C041 U5EJS0 A0A1Q3FB94 A0A182INN9 A0A1Y1NE10 A0A182Q654 A0A023EHX7 A0A336LYR2 A0A1I8PHQ7 A0A182VWM7 A0A182MPC4 A0A182PT24 A0A1A9W9I2 A0A182T2C5 A0A182XE41 A0A182LCG0 Q7QBE9 A0A182HJ82 A0A1W4XJU4 A0A2J7PNV0 A0A1B6JHE2 A0A182UEW9 A0A1I8MFU6 A0A182VJE5 A0A182JWB2 W8CA24 A0A1B6MK95 A0A034WM14 A0A1L8DVQ2 A0A0A1X017 A0A0K8UCR6 J3JUP3 A0A151X4U6 A0A067R5Z6 K7J9N1 A0A151IFX5 A0A1L8DVP9 F4WRF1 A0A182N510 A0A195BFC3 A0A158NUL0 E9IRW3 A0A1B0DAA4 A0A1B0CP48 A0A026W605 A0A195FK22 A0A151JQB5 E2AFV6 A0A182FKU6 A0A1B6DNE3 T1I235 A0A0M4EME0 A0A069DPM6 A0A232EX70 A0A0K8TRF4 A0A0N0BH82 A0A224XZU2 A0A2R7VZC9 B4K8L6 B4JEZ4 T1H6K3 A0A0V0G923 A0A3B0K5M0 A0A154PTF2 A0A2A3EMX1 B3LW75 A0A170ZI05 B4N9V6 A0A182YHI2 B4M3X2 A0A0P4VII8 A0A182RC47 Q299N8 B4QU02 B4PLZ8 A0A1J1J4F0 B3NZ35 B4G5H3
A0A2H1WP47 A0A0L7KG57 A0A3S2NA46 A0A1A9YDQ5 B0X2Y3 A0A023EJD9 Q17Q11 A0A1A9UTG7 A0A1B0G195 A0A1B0B321 A0A2M4ANM6 W5JHI4 A0A1A9ZSR9 T1E7Y6 A0A2M4C041 U5EJS0 A0A1Q3FB94 A0A182INN9 A0A1Y1NE10 A0A182Q654 A0A023EHX7 A0A336LYR2 A0A1I8PHQ7 A0A182VWM7 A0A182MPC4 A0A182PT24 A0A1A9W9I2 A0A182T2C5 A0A182XE41 A0A182LCG0 Q7QBE9 A0A182HJ82 A0A1W4XJU4 A0A2J7PNV0 A0A1B6JHE2 A0A182UEW9 A0A1I8MFU6 A0A182VJE5 A0A182JWB2 W8CA24 A0A1B6MK95 A0A034WM14 A0A1L8DVQ2 A0A0A1X017 A0A0K8UCR6 J3JUP3 A0A151X4U6 A0A067R5Z6 K7J9N1 A0A151IFX5 A0A1L8DVP9 F4WRF1 A0A182N510 A0A195BFC3 A0A158NUL0 E9IRW3 A0A1B0DAA4 A0A1B0CP48 A0A026W605 A0A195FK22 A0A151JQB5 E2AFV6 A0A182FKU6 A0A1B6DNE3 T1I235 A0A0M4EME0 A0A069DPM6 A0A232EX70 A0A0K8TRF4 A0A0N0BH82 A0A224XZU2 A0A2R7VZC9 B4K8L6 B4JEZ4 T1H6K3 A0A0V0G923 A0A3B0K5M0 A0A154PTF2 A0A2A3EMX1 B3LW75 A0A170ZI05 B4N9V6 A0A182YHI2 B4M3X2 A0A0P4VII8 A0A182RC47 Q299N8 B4QU02 B4PLZ8 A0A1J1J4F0 B3NZ35 B4G5H3
Pubmed
19121390
26354079
22118469
26227816
24945155
17510324
+ More
20920257 23761445 28004739 20966253 12364791 14747013 17210077 25315136 24495485 25348373 25830018 22516182 23537049 24845553 20075255 21719571 21347285 21282665 24508170 30249741 20798317 26334808 28648823 26369729 17994087 18057021 25244985 27129103 15632085 17550304
20920257 23761445 28004739 20966253 12364791 14747013 17210077 25315136 24495485 25348373 25830018 22516182 23537049 24845553 20075255 21719571 21347285 21282665 24508170 30249741 20798317 26334808 28648823 26369729 17994087 18057021 25244985 27129103 15632085 17550304
EMBL
BABH01022712
GDQN01004673
JAT86381.1
KQ460655
KPJ12964.1
AGBW02008320
+ More
OWR53732.1 KQ459388 KPJ01129.1 NWSH01001037 PCG72991.1 ODYU01010008 SOQ54808.1 JTDY01009900 KOB62303.1 RSAL01000158 RVE45552.1 DS232303 EDS39485.1 GAPW01004511 JAC09087.1 CH477188 EAT48783.1 CCAG010001934 JXJN01007759 GGFK01009075 MBW42396.1 ADMH02001488 ETN62370.1 GAMD01002652 JAA98938.1 GGFJ01009483 MBW58624.1 GANO01002131 JAB57740.1 GFDL01010190 JAV24855.1 GEZM01010735 JAV93867.1 AXCN02000180 GAPW01004921 JAC08677.1 UFQT01000045 SSX18808.1 AXCM01020305 AAAB01008879 EAA08395.3 APCN01002131 NEVH01023289 PNF17996.1 GECU01009072 JAS98634.1 GAMC01001654 JAC04902.1 GEBQ01012006 GEBQ01009827 GEBQ01003636 JAT27971.1 JAT30150.1 JAT36341.1 GAKP01004139 JAC54813.1 GFDF01003578 JAV10506.1 GBXI01010066 JAD04226.1 GDHF01033189 GDHF01027852 JAI19125.1 JAI24462.1 APGK01022687 BT126958 KB740442 KB631692 AEE61920.1 ENN80642.1 ERL85404.1 KQ982540 KYQ55397.1 KK852680 KDR18616.1 AAZX01002711 KQ977771 KYN00030.1 GFDF01003577 JAV10507.1 GL888285 EGI63222.1 KQ976504 KYM82870.1 ADTU01026592 GL765283 EFZ16650.1 AJVK01028951 AJWK01021422 KK107390 QOIP01000007 EZA51438.1 RLU20932.1 KQ981522 KYN40612.1 KQ978660 KYN29388.1 GL439158 EFN67705.1 GEDC01010106 JAS27192.1 ACPB03000298 CP012526 ALC46602.1 GBGD01003248 JAC85641.1 NNAY01001792 OXU22908.1 GDAI01000890 JAI16713.1 KQ435759 KOX75672.1 GFTR01002807 JAW13619.1 KK854191 PTY12854.1 CH933806 EDW14415.1 KRG01004.1 CH916369 EDV93275.1 GECL01001743 JAP04381.1 OUUW01000013 SPP87972.1 KQ435095 KZC14628.1 KZ288217 PBC32391.1 CH902617 EDV42653.1 GEMB01002179 JAS01001.1 CH964232 EDW81711.1 CH940652 EDW59333.1 GDKW01002873 JAI53722.1 CM000070 EAL27665.1 CM000364 EDX12425.1 CM000160 EDW95930.1 EDW95932.1 CVRI01000070 CRL07275.1 CH954181 EDV48577.1 CH479179 EDW24839.1
OWR53732.1 KQ459388 KPJ01129.1 NWSH01001037 PCG72991.1 ODYU01010008 SOQ54808.1 JTDY01009900 KOB62303.1 RSAL01000158 RVE45552.1 DS232303 EDS39485.1 GAPW01004511 JAC09087.1 CH477188 EAT48783.1 CCAG010001934 JXJN01007759 GGFK01009075 MBW42396.1 ADMH02001488 ETN62370.1 GAMD01002652 JAA98938.1 GGFJ01009483 MBW58624.1 GANO01002131 JAB57740.1 GFDL01010190 JAV24855.1 GEZM01010735 JAV93867.1 AXCN02000180 GAPW01004921 JAC08677.1 UFQT01000045 SSX18808.1 AXCM01020305 AAAB01008879 EAA08395.3 APCN01002131 NEVH01023289 PNF17996.1 GECU01009072 JAS98634.1 GAMC01001654 JAC04902.1 GEBQ01012006 GEBQ01009827 GEBQ01003636 JAT27971.1 JAT30150.1 JAT36341.1 GAKP01004139 JAC54813.1 GFDF01003578 JAV10506.1 GBXI01010066 JAD04226.1 GDHF01033189 GDHF01027852 JAI19125.1 JAI24462.1 APGK01022687 BT126958 KB740442 KB631692 AEE61920.1 ENN80642.1 ERL85404.1 KQ982540 KYQ55397.1 KK852680 KDR18616.1 AAZX01002711 KQ977771 KYN00030.1 GFDF01003577 JAV10507.1 GL888285 EGI63222.1 KQ976504 KYM82870.1 ADTU01026592 GL765283 EFZ16650.1 AJVK01028951 AJWK01021422 KK107390 QOIP01000007 EZA51438.1 RLU20932.1 KQ981522 KYN40612.1 KQ978660 KYN29388.1 GL439158 EFN67705.1 GEDC01010106 JAS27192.1 ACPB03000298 CP012526 ALC46602.1 GBGD01003248 JAC85641.1 NNAY01001792 OXU22908.1 GDAI01000890 JAI16713.1 KQ435759 KOX75672.1 GFTR01002807 JAW13619.1 KK854191 PTY12854.1 CH933806 EDW14415.1 KRG01004.1 CH916369 EDV93275.1 GECL01001743 JAP04381.1 OUUW01000013 SPP87972.1 KQ435095 KZC14628.1 KZ288217 PBC32391.1 CH902617 EDV42653.1 GEMB01002179 JAS01001.1 CH964232 EDW81711.1 CH940652 EDW59333.1 GDKW01002873 JAI53722.1 CM000070 EAL27665.1 CM000364 EDX12425.1 CM000160 EDW95930.1 EDW95932.1 CVRI01000070 CRL07275.1 CH954181 EDV48577.1 CH479179 EDW24839.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000218220
UP000037510
+ More
UP000283053 UP000092443 UP000002320 UP000008820 UP000078200 UP000092444 UP000092460 UP000000673 UP000092445 UP000075880 UP000075886 UP000095300 UP000075920 UP000075883 UP000075885 UP000091820 UP000075901 UP000076407 UP000075882 UP000007062 UP000075840 UP000192223 UP000235965 UP000075902 UP000095301 UP000075903 UP000075881 UP000019118 UP000030742 UP000075809 UP000027135 UP000002358 UP000078542 UP000007755 UP000075884 UP000078540 UP000005205 UP000092462 UP000092461 UP000053097 UP000279307 UP000078541 UP000078492 UP000000311 UP000069272 UP000015103 UP000092553 UP000215335 UP000053105 UP000009192 UP000001070 UP000015102 UP000268350 UP000076502 UP000242457 UP000007801 UP000007798 UP000076408 UP000008792 UP000075900 UP000001819 UP000000304 UP000002282 UP000183832 UP000008711 UP000008744
UP000283053 UP000092443 UP000002320 UP000008820 UP000078200 UP000092444 UP000092460 UP000000673 UP000092445 UP000075880 UP000075886 UP000095300 UP000075920 UP000075883 UP000075885 UP000091820 UP000075901 UP000076407 UP000075882 UP000007062 UP000075840 UP000192223 UP000235965 UP000075902 UP000095301 UP000075903 UP000075881 UP000019118 UP000030742 UP000075809 UP000027135 UP000002358 UP000078542 UP000007755 UP000075884 UP000078540 UP000005205 UP000092462 UP000092461 UP000053097 UP000279307 UP000078541 UP000078492 UP000000311 UP000069272 UP000015103 UP000092553 UP000215335 UP000053105 UP000009192 UP000001070 UP000015102 UP000268350 UP000076502 UP000242457 UP000007801 UP000007798 UP000076408 UP000008792 UP000075900 UP000001819 UP000000304 UP000002282 UP000183832 UP000008711 UP000008744
PRIDE
Interpro
IPR039175
TIM22
+ More
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR027417 P-loop_NTPase
IPR037213 Run_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR004012 Run_dom
IPR037745 SGSM1/2
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR027417 P-loop_NTPase
IPR037213 Run_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR004012 Run_dom
IPR037745 SGSM1/2
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
ProteinModelPortal
H9JUN0
A0A1E1WHR2
A0A0N0PC46
A0A212FJ28
A0A0N0P9V2
A0A2A4JLY3
+ More
A0A2H1WP47 A0A0L7KG57 A0A3S2NA46 A0A1A9YDQ5 B0X2Y3 A0A023EJD9 Q17Q11 A0A1A9UTG7 A0A1B0G195 A0A1B0B321 A0A2M4ANM6 W5JHI4 A0A1A9ZSR9 T1E7Y6 A0A2M4C041 U5EJS0 A0A1Q3FB94 A0A182INN9 A0A1Y1NE10 A0A182Q654 A0A023EHX7 A0A336LYR2 A0A1I8PHQ7 A0A182VWM7 A0A182MPC4 A0A182PT24 A0A1A9W9I2 A0A182T2C5 A0A182XE41 A0A182LCG0 Q7QBE9 A0A182HJ82 A0A1W4XJU4 A0A2J7PNV0 A0A1B6JHE2 A0A182UEW9 A0A1I8MFU6 A0A182VJE5 A0A182JWB2 W8CA24 A0A1B6MK95 A0A034WM14 A0A1L8DVQ2 A0A0A1X017 A0A0K8UCR6 J3JUP3 A0A151X4U6 A0A067R5Z6 K7J9N1 A0A151IFX5 A0A1L8DVP9 F4WRF1 A0A182N510 A0A195BFC3 A0A158NUL0 E9IRW3 A0A1B0DAA4 A0A1B0CP48 A0A026W605 A0A195FK22 A0A151JQB5 E2AFV6 A0A182FKU6 A0A1B6DNE3 T1I235 A0A0M4EME0 A0A069DPM6 A0A232EX70 A0A0K8TRF4 A0A0N0BH82 A0A224XZU2 A0A2R7VZC9 B4K8L6 B4JEZ4 T1H6K3 A0A0V0G923 A0A3B0K5M0 A0A154PTF2 A0A2A3EMX1 B3LW75 A0A170ZI05 B4N9V6 A0A182YHI2 B4M3X2 A0A0P4VII8 A0A182RC47 Q299N8 B4QU02 B4PLZ8 A0A1J1J4F0 B3NZ35 B4G5H3
A0A2H1WP47 A0A0L7KG57 A0A3S2NA46 A0A1A9YDQ5 B0X2Y3 A0A023EJD9 Q17Q11 A0A1A9UTG7 A0A1B0G195 A0A1B0B321 A0A2M4ANM6 W5JHI4 A0A1A9ZSR9 T1E7Y6 A0A2M4C041 U5EJS0 A0A1Q3FB94 A0A182INN9 A0A1Y1NE10 A0A182Q654 A0A023EHX7 A0A336LYR2 A0A1I8PHQ7 A0A182VWM7 A0A182MPC4 A0A182PT24 A0A1A9W9I2 A0A182T2C5 A0A182XE41 A0A182LCG0 Q7QBE9 A0A182HJ82 A0A1W4XJU4 A0A2J7PNV0 A0A1B6JHE2 A0A182UEW9 A0A1I8MFU6 A0A182VJE5 A0A182JWB2 W8CA24 A0A1B6MK95 A0A034WM14 A0A1L8DVQ2 A0A0A1X017 A0A0K8UCR6 J3JUP3 A0A151X4U6 A0A067R5Z6 K7J9N1 A0A151IFX5 A0A1L8DVP9 F4WRF1 A0A182N510 A0A195BFC3 A0A158NUL0 E9IRW3 A0A1B0DAA4 A0A1B0CP48 A0A026W605 A0A195FK22 A0A151JQB5 E2AFV6 A0A182FKU6 A0A1B6DNE3 T1I235 A0A0M4EME0 A0A069DPM6 A0A232EX70 A0A0K8TRF4 A0A0N0BH82 A0A224XZU2 A0A2R7VZC9 B4K8L6 B4JEZ4 T1H6K3 A0A0V0G923 A0A3B0K5M0 A0A154PTF2 A0A2A3EMX1 B3LW75 A0A170ZI05 B4N9V6 A0A182YHI2 B4M3X2 A0A0P4VII8 A0A182RC47 Q299N8 B4QU02 B4PLZ8 A0A1J1J4F0 B3NZ35 B4G5H3
Ontologies
PANTHER
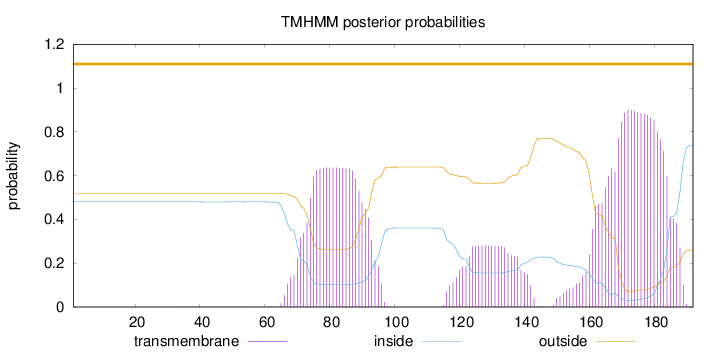
Topology
Length:
192
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
39.78794
Exp number, first 60 AAs:
0.00995
Total prob of N-in:
0.48030
outside
1 - 192
Population Genetic Test Statistics
Pi
161.526747
Theta
153.594651
Tajima's D
0.272459
CLR
0
CSRT
0.446577671116444
Interpretation
Uncertain
