Pre Gene Modal
BGIBMGA013245
Annotation
PREDICTED:_ectonucleoside_triphosphate_diphosphohydrolase_5_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.01
Sequence
CDS
ATGGAAATTCGTCAAAGAAAGTTAAAAGATGATGTCCCCTCAACCCAAACTCATATTCGAAGCAGAGGTATAAAACAGCCAAGGCGTCTGAAGAAAGTATTCATTCTTTTGATTGTTTTGGCACTAGTCATAACATATTTATTGGGTCTGTTCGCCGGCCAATCACAATGGCTCGATAGCATCGCCAAGACCCTCGGCTACCAGAGCGAGTACCATTACGCTGTGATAATCGACGCTGGTTCGTCGGGAAGTCGGGTACTCGCTTATAAATTCAGAGTTCCTTTTACAGTATTCAGTCAAGCCAACCTAGACTTAGTTCAAGAGCACTTTGAGCAGTCCAAACCTGGGCTGTCATCGTTTGCTGATGATCCAGCAAAGGGGGCAGAGACAATAGTGCAATTAATAAAAAAGGCGGAATACTTGATACCGTTGGAGAAGAGGAAATCGACCCCCCTCATAGTACGAGCCACGGCCGGCCTGAGGCTGTTGCCTGCGGACCGCGCCGAGCAGCTCCTCGCACACGTCAAGAAGGCCATTTCCGTTCTGGGCTACGATGTTGGTCCGAACAGCGTGGACATCATGGACGGCGCGGACGAGGGCATCTACATATGGTACACGGTCAACTTGCTGCATAATCTTATCGAGGGCGAGACGATGGCAGCCCTGGACCTGGGCGGCGGCTCCACGCAGATCACGTTCCAGCTGGCGGAGGGCGAGCGGGCGCGCTACCCGCCGCGCGACCTGCACGTGGTGCCCGCGGGCAGCAACATCACGCTCTACACGCACAGCTACCTGGGTCTGGGGCTGCTGGCGGCTCGCTACGGGGTCCTGCTGCAGGAGGCCGGGGGGAACGACACGGGAGAGTTCAGCTCCGTCTGCGTGGAGCCCATCATCAAGGACGAGCCCTGGGTCTACGCCAACAAGCAGTACCGCGTCAGCGGAGCGCCCCGCAAGCCGGGCACGAAGCGCAAGGCGGCGTTCGAGGCGTGCCTGGAGGCGGTGAAGCAGTACCTGGACGGCGCGCTGGGCGGCGGCGGCGCGCCGGGCCCGCGCGGCAGCCTGGCCGCCATGAGCTTCTTCTACGACGTCGCCGCCGACGCCGGGATCATTGACGTGGCGCGCGGCGGCACGGGCAGCGTGGCGCAGTACTCGCGCGCGGCGCAGCGGGCGTGCTCGGCGCACAACGTGGAGCAGCCCTGGGCGTGCCTGGACCTGGTGTACGTGCTGGCGCTGCTGCACCACGCCTACAGGGTGCCGACCGAACAGCCAGTATATCTGTTCAAGAAAGTGAACGGCCACGAAGTATCCTGGGCCCTCGGCTTGGCGTACTCCACGGTGATGAACAGAATAACTACAAAATAA
Protein
MEIRQRKLKDDVPSTQTHIRSRGIKQPRRLKKVFILLIVLALVITYLLGLFAGQSQWLDSIAKTLGYQSEYHYAVIIDAGSSGSRVLAYKFRVPFTVFSQANLDLVQEHFEQSKPGLSSFADDPAKGAETIVQLIKKAEYLIPLEKRKSTPLIVRATAGLRLLPADRAEQLLAHVKKAISVLGYDVGPNSVDIMDGADEGIYIWYTVNLLHNLIEGETMAALDLGGGSTQITFQLAEGERARYPPRDLHVVPAGSNITLYTHSYLGLGLLAARYGVLLQEAGGNDTGEFSSVCVEPIIKDEPWVYANKQYRVSGAPRKPGTKRKAAFEACLEAVKQYLDGALGGGGAPGPRGSLAAMSFFYDVAADAGIIDVARGGTGSVAQYSRAAQRACSAHNVEQPWACLDLVYVLALLHHAYRVPTEQPVYLFKKVNGHEVSWALGLAYSTVMNRITTK
Summary
Similarity
Belongs to the GDA1/CD39 NTPase family.
Uniprot
H9JUN2
A0A437B4X1
A0A2H1W7K9
A0A0N1I8U6
A0A1E1WB04
A0A034WVD9
+ More
B4N7P3 W8B8B8 A0A034WSW9 A0A1L8EDF4 T1PMJ7 A0A3B0K8L2 A0A1L8EDC6 A0A0K8ULY0 A0A0Q9WUP4 A0A1I8NP57 A0A3B0KBF5 A0A0M4E6L4 A0A1I8NP55 A0A1I8M8Z5 A0A0P8ZEA8 B3MLB5 A0A0K8TW41 A0A1I8NP56 A0A1I8M906 A0A0P8XUK9 A0A0A1X0N7 B4JC75 A0A3B0KFH7 B4GSB1 A0A0Q9WB62 A0A0Q5W8V6 A0A3B0K394 A0A0R3NV52 Q9VQI8 B3NAM6 O76268 B4M9I5 Q29KG3 A0A0Q5W8Y7 A0A0N8NZ07 A0A0J9QUM8 B4Q8M9 A0A0J9TEB3 A0A1W4V7W1 B4KH04 A0A0Q9XDW7 A0A0R3NZW3 M9PBV2 A0A0Q9WAA7 A0A1W4UU63 A0A0Q5WKM5 B4I2S8 A0A0Q9XHX9 D5AEL4 A0A0R1DL53 B4NXA7 A0A0R1DIR3 A0A0J9QUS2 A0A1W4UUK4 A0A0L0BVF1 A0A2J7R2I1 A0A2J7R2H4 A0A0P4WLA2 A0A2J7R2H9 A0A0P4WHT5 A0A0R1DIU1 A0A182RIP3 A0A182M4V9 A0A182VUG6 A0A182IMJ0 A0A182NU47 A0A182QRL3 A0A182WW38 A0A182VGP0 A0A182U497 A0A1A9Y4Z4 A0A1B0AWW5 A0A182HGH4 A0A084WG01 A0A336M055 A0A1S4H0M9 Q7PVI8 U5EHH5 A0A182L6D1 A0A1J1IN73 A0A182PE87 A0A1J1IL86 A0A182F3K1 A0A3F2YPT4 A0A1S4FBA9 Q179G0 A0A2M4ACM5 A0A1B0D5B7 A0A2M4ACE3 A0A2M3Z2D0 W5JGB9
B4N7P3 W8B8B8 A0A034WSW9 A0A1L8EDF4 T1PMJ7 A0A3B0K8L2 A0A1L8EDC6 A0A0K8ULY0 A0A0Q9WUP4 A0A1I8NP57 A0A3B0KBF5 A0A0M4E6L4 A0A1I8NP55 A0A1I8M8Z5 A0A0P8ZEA8 B3MLB5 A0A0K8TW41 A0A1I8NP56 A0A1I8M906 A0A0P8XUK9 A0A0A1X0N7 B4JC75 A0A3B0KFH7 B4GSB1 A0A0Q9WB62 A0A0Q5W8V6 A0A3B0K394 A0A0R3NV52 Q9VQI8 B3NAM6 O76268 B4M9I5 Q29KG3 A0A0Q5W8Y7 A0A0N8NZ07 A0A0J9QUM8 B4Q8M9 A0A0J9TEB3 A0A1W4V7W1 B4KH04 A0A0Q9XDW7 A0A0R3NZW3 M9PBV2 A0A0Q9WAA7 A0A1W4UU63 A0A0Q5WKM5 B4I2S8 A0A0Q9XHX9 D5AEL4 A0A0R1DL53 B4NXA7 A0A0R1DIR3 A0A0J9QUS2 A0A1W4UUK4 A0A0L0BVF1 A0A2J7R2I1 A0A2J7R2H4 A0A0P4WLA2 A0A2J7R2H9 A0A0P4WHT5 A0A0R1DIU1 A0A182RIP3 A0A182M4V9 A0A182VUG6 A0A182IMJ0 A0A182NU47 A0A182QRL3 A0A182WW38 A0A182VGP0 A0A182U497 A0A1A9Y4Z4 A0A1B0AWW5 A0A182HGH4 A0A084WG01 A0A336M055 A0A1S4H0M9 Q7PVI8 U5EHH5 A0A182L6D1 A0A1J1IN73 A0A182PE87 A0A1J1IL86 A0A182F3K1 A0A3F2YPT4 A0A1S4FBA9 Q179G0 A0A2M4ACM5 A0A1B0D5B7 A0A2M4ACE3 A0A2M3Z2D0 W5JGB9
Pubmed
EMBL
BABH01022720
RSAL01000158
RVE45557.1
ODYU01006846
SOQ49060.1
KQ460655
+ More
KPJ12969.1 GDQN01006882 JAT84172.1 GAKP01001234 JAC57718.1 CH964214 EDW80382.2 GAMC01013232 JAB93323.1 GAKP01001233 JAC57719.1 GFDG01002039 JAV16760.1 KA649977 AFP64606.1 OUUW01000006 SPP82389.1 GFDG01002059 JAV16740.1 GDHF01024612 JAI27702.1 KRF99272.1 SPP82391.1 CP012523 ALC40008.1 CH902620 KPU72995.1 EDV30704.1 GDHF01033806 JAI18508.1 KPU72997.1 GBXI01009997 JAD04295.1 CH916368 EDW03088.1 SPP82388.1 CH479189 EDW25658.1 CH940654 KRF77982.1 CH954177 KQS70016.1 SPP82390.1 CH379061 KRT04812.1 AE014134 BT133416 AAF51181.1 AFH58713.1 EDV57549.2 AF041048 AY061134 EU852293 AAC39133.1 AAF51182.1 AAL28682.1 ACF58165.1 EDW57861.1 EAL33212.2 KRT04811.1 KQS70017.1 KPU72996.1 CM002910 KMY87777.1 CM000361 EDX03546.1 KMY87774.1 KMY87775.1 CH933807 EDW12215.1 KRG03156.1 KRT04813.1 AGB92509.1 KRF77983.1 KQS70018.1 CH480820 EDW54073.1 KRG03155.1 BT124751 ADF28792.1 CM000157 KRJ97165.1 EDW87464.2 KRJ97164.1 KMY87776.1 JRES01001277 KNC24017.1 NEVH01007830 PNF35032.1 PNF35035.1 GDRN01039809 JAI67181.1 PNF35031.1 GDRN01039808 JAI67182.1 KRJ97163.1 AXCM01000998 AXCN02000773 JXJN01004959 APCN01004232 ATLV01023416 KE525343 KFB49145.1 UFQT01000369 SSX23615.1 AAAB01008984 EAA14912.3 GANO01003020 JAB56851.1 CVRI01000055 CRL01004.1 CRL01005.1 CH477353 EAT42846.1 GGFK01005212 MBW38533.1 AJVK01011729 GGFK01005144 MBW38465.1 GGFM01001931 MBW22682.1 ADMH02001595 ETN61925.1
KPJ12969.1 GDQN01006882 JAT84172.1 GAKP01001234 JAC57718.1 CH964214 EDW80382.2 GAMC01013232 JAB93323.1 GAKP01001233 JAC57719.1 GFDG01002039 JAV16760.1 KA649977 AFP64606.1 OUUW01000006 SPP82389.1 GFDG01002059 JAV16740.1 GDHF01024612 JAI27702.1 KRF99272.1 SPP82391.1 CP012523 ALC40008.1 CH902620 KPU72995.1 EDV30704.1 GDHF01033806 JAI18508.1 KPU72997.1 GBXI01009997 JAD04295.1 CH916368 EDW03088.1 SPP82388.1 CH479189 EDW25658.1 CH940654 KRF77982.1 CH954177 KQS70016.1 SPP82390.1 CH379061 KRT04812.1 AE014134 BT133416 AAF51181.1 AFH58713.1 EDV57549.2 AF041048 AY061134 EU852293 AAC39133.1 AAF51182.1 AAL28682.1 ACF58165.1 EDW57861.1 EAL33212.2 KRT04811.1 KQS70017.1 KPU72996.1 CM002910 KMY87777.1 CM000361 EDX03546.1 KMY87774.1 KMY87775.1 CH933807 EDW12215.1 KRG03156.1 KRT04813.1 AGB92509.1 KRF77983.1 KQS70018.1 CH480820 EDW54073.1 KRG03155.1 BT124751 ADF28792.1 CM000157 KRJ97165.1 EDW87464.2 KRJ97164.1 KMY87776.1 JRES01001277 KNC24017.1 NEVH01007830 PNF35032.1 PNF35035.1 GDRN01039809 JAI67181.1 PNF35031.1 GDRN01039808 JAI67182.1 KRJ97163.1 AXCM01000998 AXCN02000773 JXJN01004959 APCN01004232 ATLV01023416 KE525343 KFB49145.1 UFQT01000369 SSX23615.1 AAAB01008984 EAA14912.3 GANO01003020 JAB56851.1 CVRI01000055 CRL01004.1 CRL01005.1 CH477353 EAT42846.1 GGFK01005212 MBW38533.1 AJVK01011729 GGFK01005144 MBW38465.1 GGFM01001931 MBW22682.1 ADMH02001595 ETN61925.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007798
UP000268350
UP000095300
+ More
UP000092553 UP000095301 UP000007801 UP000001070 UP000008744 UP000008792 UP000008711 UP000001819 UP000000803 UP000000304 UP000192221 UP000009192 UP000001292 UP000002282 UP000037069 UP000235965 UP000075900 UP000075883 UP000075920 UP000075880 UP000075884 UP000075886 UP000076407 UP000075903 UP000075902 UP000092443 UP000092460 UP000075840 UP000030765 UP000007062 UP000075882 UP000183832 UP000075885 UP000069272 UP000008820 UP000092462 UP000000673
UP000092553 UP000095301 UP000007801 UP000001070 UP000008744 UP000008792 UP000008711 UP000001819 UP000000803 UP000000304 UP000192221 UP000009192 UP000001292 UP000002282 UP000037069 UP000235965 UP000075900 UP000075883 UP000075920 UP000075880 UP000075884 UP000075886 UP000076407 UP000075903 UP000075902 UP000092443 UP000092460 UP000075840 UP000030765 UP000007062 UP000075882 UP000183832 UP000075885 UP000069272 UP000008820 UP000092462 UP000000673
Pfam
PF01150 GDA1_CD39
Interpro
IPR000407
GDA1_CD39_NTPase
ProteinModelPortal
H9JUN2
A0A437B4X1
A0A2H1W7K9
A0A0N1I8U6
A0A1E1WB04
A0A034WVD9
+ More
B4N7P3 W8B8B8 A0A034WSW9 A0A1L8EDF4 T1PMJ7 A0A3B0K8L2 A0A1L8EDC6 A0A0K8ULY0 A0A0Q9WUP4 A0A1I8NP57 A0A3B0KBF5 A0A0M4E6L4 A0A1I8NP55 A0A1I8M8Z5 A0A0P8ZEA8 B3MLB5 A0A0K8TW41 A0A1I8NP56 A0A1I8M906 A0A0P8XUK9 A0A0A1X0N7 B4JC75 A0A3B0KFH7 B4GSB1 A0A0Q9WB62 A0A0Q5W8V6 A0A3B0K394 A0A0R3NV52 Q9VQI8 B3NAM6 O76268 B4M9I5 Q29KG3 A0A0Q5W8Y7 A0A0N8NZ07 A0A0J9QUM8 B4Q8M9 A0A0J9TEB3 A0A1W4V7W1 B4KH04 A0A0Q9XDW7 A0A0R3NZW3 M9PBV2 A0A0Q9WAA7 A0A1W4UU63 A0A0Q5WKM5 B4I2S8 A0A0Q9XHX9 D5AEL4 A0A0R1DL53 B4NXA7 A0A0R1DIR3 A0A0J9QUS2 A0A1W4UUK4 A0A0L0BVF1 A0A2J7R2I1 A0A2J7R2H4 A0A0P4WLA2 A0A2J7R2H9 A0A0P4WHT5 A0A0R1DIU1 A0A182RIP3 A0A182M4V9 A0A182VUG6 A0A182IMJ0 A0A182NU47 A0A182QRL3 A0A182WW38 A0A182VGP0 A0A182U497 A0A1A9Y4Z4 A0A1B0AWW5 A0A182HGH4 A0A084WG01 A0A336M055 A0A1S4H0M9 Q7PVI8 U5EHH5 A0A182L6D1 A0A1J1IN73 A0A182PE87 A0A1J1IL86 A0A182F3K1 A0A3F2YPT4 A0A1S4FBA9 Q179G0 A0A2M4ACM5 A0A1B0D5B7 A0A2M4ACE3 A0A2M3Z2D0 W5JGB9
B4N7P3 W8B8B8 A0A034WSW9 A0A1L8EDF4 T1PMJ7 A0A3B0K8L2 A0A1L8EDC6 A0A0K8ULY0 A0A0Q9WUP4 A0A1I8NP57 A0A3B0KBF5 A0A0M4E6L4 A0A1I8NP55 A0A1I8M8Z5 A0A0P8ZEA8 B3MLB5 A0A0K8TW41 A0A1I8NP56 A0A1I8M906 A0A0P8XUK9 A0A0A1X0N7 B4JC75 A0A3B0KFH7 B4GSB1 A0A0Q9WB62 A0A0Q5W8V6 A0A3B0K394 A0A0R3NV52 Q9VQI8 B3NAM6 O76268 B4M9I5 Q29KG3 A0A0Q5W8Y7 A0A0N8NZ07 A0A0J9QUM8 B4Q8M9 A0A0J9TEB3 A0A1W4V7W1 B4KH04 A0A0Q9XDW7 A0A0R3NZW3 M9PBV2 A0A0Q9WAA7 A0A1W4UU63 A0A0Q5WKM5 B4I2S8 A0A0Q9XHX9 D5AEL4 A0A0R1DL53 B4NXA7 A0A0R1DIR3 A0A0J9QUS2 A0A1W4UUK4 A0A0L0BVF1 A0A2J7R2I1 A0A2J7R2H4 A0A0P4WLA2 A0A2J7R2H9 A0A0P4WHT5 A0A0R1DIU1 A0A182RIP3 A0A182M4V9 A0A182VUG6 A0A182IMJ0 A0A182NU47 A0A182QRL3 A0A182WW38 A0A182VGP0 A0A182U497 A0A1A9Y4Z4 A0A1B0AWW5 A0A182HGH4 A0A084WG01 A0A336M055 A0A1S4H0M9 Q7PVI8 U5EHH5 A0A182L6D1 A0A1J1IN73 A0A182PE87 A0A1J1IL86 A0A182F3K1 A0A3F2YPT4 A0A1S4FBA9 Q179G0 A0A2M4ACM5 A0A1B0D5B7 A0A2M4ACE3 A0A2M3Z2D0 W5JGB9
PDB
5U7W
E-value=1.06042e-28,
Score=316
Ontologies
PATHWAY
GO
PANTHER
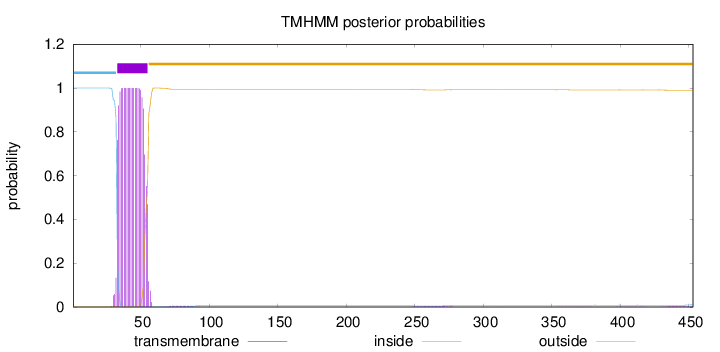
Topology
Length:
453
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.96241
Exp number, first 60 AAs:
21.69466
Total prob of N-in:
0.99975
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 453
Population Genetic Test Statistics
Pi
316.915745
Theta
178.097764
Tajima's D
2.23664
CLR
0.246027
CSRT
0.910754462276886
Interpretation
Uncertain
