Gene
KWMTBOMO09922
Pre Gene Modal
BGIBMGA013171
Annotation
PREDICTED:_phosphorylase_b_kinase_gamma_catalytic_chain?_skeletal_muscle/heart_isoform_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.632
Sequence
CDS
ATGGCTAAGGAAGAAGATGATGATTTACCAGACAAAGATGCAGCCAAGGAATTTTACGCAAAATATGAACCCAAAGAAATCATAGGCAGAGGCATAAGTTCTACTGTGCGAAGATGCATAGAGAAGGAAACGGGACGAGAATATGCAGCCAAGATCATAGACCTAAGTCAGGAATCACAAGATGGAGTGGACACACACACAATGAGAGATGCCACTCGTCAAGAAATACATATATTGAGGATGGTCTCTGGACACCCATACATTATTGAACTTCAAGATGTGTTTGAATCAGAGACATTCATATTCTTAGTATTTGAGCTGTGCAAGAATGGTGAATTATTTGACTATTTGACATCTGTAGTTACACTCTCTGAAAAGAAAACTAGGTATATAATGAGACAAGTGCTCGAAGGCGTGAGACACGTGCACAGCAAGAACATCGTTCACAGAGACCTGAAGCCGGAGAACATTCTGCTGGACGATCAGCTCAACGTCAAGATCACAGACTTCGGCTTCGCGAAGGTGCTCAAAGACGGGGAGAGACTGTTCGAATTGTGCGGCACGCCCGGCTACCTTGCCCCGGAGACGCTGAAGGCGAATATGTTCGAAGACGCTCCTGGCTACGGGATGGAGGTCGACTTATGGGCTTGCGGAGTCATCATGTTCACATTACTGGTGGGATGTCCTCCGTTCTGGCACCGGAAGCAGATGGTGATGCTGAGGAATATAATGGAGGGGAAATATTCTTTTACCAGCCCCGAGTGGGTGGATATATCAGACGATCCTAAAGATCTCATCCGACGGTTTCTAGTAGTGGAACCGAGTAACAGGATAGACCTGATGGACGCTTTAAATCATCCCTTCTTCAACACTGTGCTGTGGGACCAGGATATGAAGCCGCTGAAGCAGAGCTTCTCGACGCAAAGCCGCAGGATGTCTAGGATCGAGCAGCTCAGCATGGCAATGAAGTGCGGCAGCTTCAACGGGCGCGCCAAGCTGCGCGTGGCGCTGGTGACGGTGCGCGCGGCGGTGCGGCTGCGGCGCCTCCCGCACACCGCGGCGCGCCCGCTGCCCCTCGCCGACGCCGCCAGGGACCCCTACGCCGTGCGCGCGCTCAGGAAGGTGATAGACGGGTGCGCGTTCCGCGTGTACGGGCACTGGGTGAAGAAGGGCGAGGGACAGAACCGCGCCGCCCTGTTCGAGAACATGCCAAAGACCGAACTCAAGGCTCTGTACGTGAGCAACCTCAGCCGGTAG
Protein
MAKEEDDDLPDKDAAKEFYAKYEPKEIIGRGISSTVRRCIEKETGREYAAKIIDLSQESQDGVDTHTMRDATRQEIHILRMVSGHPYIIELQDVFESETFIFLVFELCKNGELFDYLTSVVTLSEKKTRYIMRQVLEGVRHVHSKNIVHRDLKPENILLDDQLNVKITDFGFAKVLKDGERLFELCGTPGYLAPETLKANMFEDAPGYGMEVDLWACGVIMFTLLVGCPPFWHRKQMVMLRNIMEGKYSFTSPEWVDISDDPKDLIRRFLVVEPSNRIDLMDALNHPFFNTVLWDQDMKPLKQSFSTQSRRMSRIEQLSMAMKCGSFNGRAKLRVALVTVRAAVRLRRLPHTAARPLPLADAARDPYAVRALRKVIDGCAFRVYGHWVKKGEGQNRAALFENMPKTELKALYVSNLSR
Summary
Similarity
Belongs to the protein kinase superfamily.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Uniprot
H9JUF8
A0A2A4IWE8
A0A437BU50
A0A194RED7
A0A212FM53
A0A194PSR1
+ More
A0A0M9ABB5 A0A0C9QVJ3 E0W3N2 A0A232EHU0 A0A1L8DKE8 A0A2A3EI29 A0A088AJT9 A0A310SFA0 U5EP90 E2ASE5 A0A1Q3FYX2 A0A084W753 A0A182XAJ9 Q7QCI6 A0A023ERV0 A0A182W0Q8 A0A3L8D7K5 A0A2M4BPE0 A0A2M4CTE8 A0A2M4A975 A0A182RQJ5 D2A4H2 A0A0L7RJX9 A0A1I8P1T5 A0A195CNT7 T1E8Y1 T1PB93 A0A1Q3FYT5 E2BJJ3 A0A2M4CSE6 A0A2M3ZH04 A0A2M4A985 A0A2M4BP87 A0A1B6HQJ5 A0A154PBV3 A0A1B6F300 A0A1W4WBI3 A0A1B6M6C1 Q961E7 B4MT15 B4MAI4 B4L681 A0A0R1EHZ8 A0A0Q5T2Z1 W8AC87 A0A3B0JWG7 Q29JA5 A0A2C9GR37 A0A0K8TL11 A0A0A1XKV0 D3TMY4 A0A2J7QK37 A0A067REZ3 A0A0K8VCD3 A0A1B6E198 U4UPH0 A0A151JYP6 A0A1A9VB09 A0A026WDA5 N6T2S8 A0A1B0CKC5 A0A1L8DKH8 A0A1Q3G4F5 A0A182QYM8 A0A182VMZ9 A0A1B6K709 A0A0P6JRY3 A0A1S3DUD7 A0A2M4CTQ9 A0A1Y1MNH7 A0A1Q3G4G1 A0A182J7P2 A0A1I8NAV9 A0A1I8P1Z7 A0A2J7QK44 A0A0R1EFD0 A0A0Q5THS5 A0A1W4VZV3 A0A0P9BUP0 T1I9G0 A0A182P3U1 A0A0Q9WES2 X2JEJ4 A0A0Q9XER1 A0A0A1WK05 A0A0P4VKY8 A0A0R3NY12 A0A2S2QMI6 A0A2H8TZN2 E9J9F9 A0A0A9VSD2 J9K8F1 A0A0A9Z894 A0A146L888
A0A0M9ABB5 A0A0C9QVJ3 E0W3N2 A0A232EHU0 A0A1L8DKE8 A0A2A3EI29 A0A088AJT9 A0A310SFA0 U5EP90 E2ASE5 A0A1Q3FYX2 A0A084W753 A0A182XAJ9 Q7QCI6 A0A023ERV0 A0A182W0Q8 A0A3L8D7K5 A0A2M4BPE0 A0A2M4CTE8 A0A2M4A975 A0A182RQJ5 D2A4H2 A0A0L7RJX9 A0A1I8P1T5 A0A195CNT7 T1E8Y1 T1PB93 A0A1Q3FYT5 E2BJJ3 A0A2M4CSE6 A0A2M3ZH04 A0A2M4A985 A0A2M4BP87 A0A1B6HQJ5 A0A154PBV3 A0A1B6F300 A0A1W4WBI3 A0A1B6M6C1 Q961E7 B4MT15 B4MAI4 B4L681 A0A0R1EHZ8 A0A0Q5T2Z1 W8AC87 A0A3B0JWG7 Q29JA5 A0A2C9GR37 A0A0K8TL11 A0A0A1XKV0 D3TMY4 A0A2J7QK37 A0A067REZ3 A0A0K8VCD3 A0A1B6E198 U4UPH0 A0A151JYP6 A0A1A9VB09 A0A026WDA5 N6T2S8 A0A1B0CKC5 A0A1L8DKH8 A0A1Q3G4F5 A0A182QYM8 A0A182VMZ9 A0A1B6K709 A0A0P6JRY3 A0A1S3DUD7 A0A2M4CTQ9 A0A1Y1MNH7 A0A1Q3G4G1 A0A182J7P2 A0A1I8NAV9 A0A1I8P1Z7 A0A2J7QK44 A0A0R1EFD0 A0A0Q5THS5 A0A1W4VZV3 A0A0P9BUP0 T1I9G0 A0A182P3U1 A0A0Q9WES2 X2JEJ4 A0A0Q9XER1 A0A0A1WK05 A0A0P4VKY8 A0A0R3NY12 A0A2S2QMI6 A0A2H8TZN2 E9J9F9 A0A0A9VSD2 J9K8F1 A0A0A9Z894 A0A146L888
Pubmed
19121390
26354079
22118469
20566863
28648823
20798317
+ More
24438588 12364791 14747013 17210077 24945155 30249741 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 18057021 17550304 24495485 15632085 23185243 26369729 25830018 20353571 24845553 23537049 24508170 26999592 28004739 27129103 21282665 25401762 26823975
24438588 12364791 14747013 17210077 24945155 30249741 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 18057021 17550304 24495485 15632085 23185243 26369729 25830018 20353571 24845553 23537049 24508170 26999592 28004739 27129103 21282665 25401762 26823975
EMBL
BABH01022742
NWSH01006418
PCG63443.1
RSAL01000008
RVE53984.1
KQ460323
+ More
KPJ15952.1 AGBW02007655 OWR54823.1 KQ459595 KPI96008.1 KQ435703 KOX80326.1 GBYB01007734 JAG77501.1 AAZO01007428 DS235882 EEB20238.1 NNAY01004439 OXU17892.1 GFDF01007146 JAV06938.1 KZ288238 PBC31358.1 KQ766578 OAD53686.1 GANO01000260 JAB59611.1 GL442298 EFN63611.1 GFDL01002312 JAV32733.1 ATLV01021109 KE525312 KFB46047.1 AAAB01008859 EAA08212.3 GAPW01001665 JAC11933.1 QOIP01000012 RLU16244.1 GGFJ01005774 MBW54915.1 GGFL01004273 MBW68451.1 GGFK01004033 MBW37354.1 KQ971344 EFA05218.1 KQ414579 KOC71139.1 KQ977481 KYN02381.1 GAMD01002358 JAA99232.1 KA645992 AFP60621.1 GFDL01002310 JAV32735.1 GL448571 EFN84150.1 GGFL01004055 MBW68233.1 GGFM01007048 MBW27799.1 GGFK01004043 MBW37364.1 GGFJ01005691 MBW54832.1 GECU01030757 JAS76949.1 KQ434869 KZC09311.1 GECZ01025117 JAS44652.1 GEBQ01008498 JAT31479.1 AY051635 AE014298 AAK93059.1 AAN09640.1 AHN59604.1 CH963851 EDW75254.1 KRF98211.1 CH940655 EDW66243.2 CH933812 EDW05877.1 CM000162 KRK06802.1 CH954180 KQS29795.1 GAMC01021090 JAB85465.1 OUUW01000003 SPP77706.1 CH379063 EAL32396.3 KRT05962.1 KRT05964.1 APCN01000266 GDAI01002767 JAI14836.1 GBXI01015900 GBXI01004361 GBXI01002666 JAC98391.1 JAD09931.1 JAD11626.1 EZ422786 ADD19062.1 NEVH01013278 PNF28950.1 KK852680 KDR18634.1 GDHF01015776 GDHF01002151 JAI36538.1 JAI50163.1 GEDC01005600 JAS31698.1 KB632404 ERL95012.1 KQ981463 KYN41500.1 KK107261 EZA53948.1 APGK01054667 KB741251 ENN71858.1 AJWK01015973 GFDF01007115 JAV06969.1 GFDL01000353 JAV34692.1 AXCN02001467 GECU01000464 JAT07243.1 GDUN01000870 JAN95049.1 GGFL01004556 MBW68734.1 GEZM01026128 JAV87223.1 GFDL01000355 JAV34690.1 PNF28951.1 KRK06803.1 KQS29796.1 CH902622 KPU75243.1 ACPB03017049 KRF82614.1 AHN59603.1 KRG07068.1 GBXI01015105 JAC99186.1 GDKW01001557 JAI55038.1 KRT05963.1 GGMS01009764 MBY78967.1 GFXV01007921 MBW19726.1 GL769304 EFZ10545.1 GBHO01044980 GBHO01044971 GBHO01041103 GBHO01002055 GBRD01017816 GBRD01011355 GBRD01011354 GBRD01011353 GDHC01015985 JAF98623.1 JAF98632.1 JAG02501.1 JAG41549.1 JAG48011.1 JAQ02644.1 ABLF02021733 GBHO01044981 GBHO01044972 GBHO01042740 GBHO01002122 JAF98622.1 JAF98631.1 JAG00864.1 JAG41482.1 GDHC01015203 JAQ03426.1
KPJ15952.1 AGBW02007655 OWR54823.1 KQ459595 KPI96008.1 KQ435703 KOX80326.1 GBYB01007734 JAG77501.1 AAZO01007428 DS235882 EEB20238.1 NNAY01004439 OXU17892.1 GFDF01007146 JAV06938.1 KZ288238 PBC31358.1 KQ766578 OAD53686.1 GANO01000260 JAB59611.1 GL442298 EFN63611.1 GFDL01002312 JAV32733.1 ATLV01021109 KE525312 KFB46047.1 AAAB01008859 EAA08212.3 GAPW01001665 JAC11933.1 QOIP01000012 RLU16244.1 GGFJ01005774 MBW54915.1 GGFL01004273 MBW68451.1 GGFK01004033 MBW37354.1 KQ971344 EFA05218.1 KQ414579 KOC71139.1 KQ977481 KYN02381.1 GAMD01002358 JAA99232.1 KA645992 AFP60621.1 GFDL01002310 JAV32735.1 GL448571 EFN84150.1 GGFL01004055 MBW68233.1 GGFM01007048 MBW27799.1 GGFK01004043 MBW37364.1 GGFJ01005691 MBW54832.1 GECU01030757 JAS76949.1 KQ434869 KZC09311.1 GECZ01025117 JAS44652.1 GEBQ01008498 JAT31479.1 AY051635 AE014298 AAK93059.1 AAN09640.1 AHN59604.1 CH963851 EDW75254.1 KRF98211.1 CH940655 EDW66243.2 CH933812 EDW05877.1 CM000162 KRK06802.1 CH954180 KQS29795.1 GAMC01021090 JAB85465.1 OUUW01000003 SPP77706.1 CH379063 EAL32396.3 KRT05962.1 KRT05964.1 APCN01000266 GDAI01002767 JAI14836.1 GBXI01015900 GBXI01004361 GBXI01002666 JAC98391.1 JAD09931.1 JAD11626.1 EZ422786 ADD19062.1 NEVH01013278 PNF28950.1 KK852680 KDR18634.1 GDHF01015776 GDHF01002151 JAI36538.1 JAI50163.1 GEDC01005600 JAS31698.1 KB632404 ERL95012.1 KQ981463 KYN41500.1 KK107261 EZA53948.1 APGK01054667 KB741251 ENN71858.1 AJWK01015973 GFDF01007115 JAV06969.1 GFDL01000353 JAV34692.1 AXCN02001467 GECU01000464 JAT07243.1 GDUN01000870 JAN95049.1 GGFL01004556 MBW68734.1 GEZM01026128 JAV87223.1 GFDL01000355 JAV34690.1 PNF28951.1 KRK06803.1 KQS29796.1 CH902622 KPU75243.1 ACPB03017049 KRF82614.1 AHN59603.1 KRG07068.1 GBXI01015105 JAC99186.1 GDKW01001557 JAI55038.1 KRT05963.1 GGMS01009764 MBY78967.1 GFXV01007921 MBW19726.1 GL769304 EFZ10545.1 GBHO01044980 GBHO01044971 GBHO01041103 GBHO01002055 GBRD01017816 GBRD01011355 GBRD01011354 GBRD01011353 GDHC01015985 JAF98623.1 JAF98632.1 JAG02501.1 JAG41549.1 JAG48011.1 JAQ02644.1 ABLF02021733 GBHO01044981 GBHO01044972 GBHO01042740 GBHO01002122 JAF98622.1 JAF98631.1 JAG00864.1 JAG41482.1 GDHC01015203 JAQ03426.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000053105 UP000009046 UP000215335 UP000242457 UP000005203 UP000000311 UP000030765 UP000076407 UP000007062 UP000075920 UP000279307 UP000075900 UP000007266 UP000053825 UP000095300 UP000078542 UP000095301 UP000008237 UP000076502 UP000192221 UP000000803 UP000007798 UP000008792 UP000009192 UP000002282 UP000008711 UP000268350 UP000001819 UP000075840 UP000235965 UP000027135 UP000030742 UP000078541 UP000078200 UP000053097 UP000019118 UP000092461 UP000075886 UP000075903 UP000079169 UP000075880 UP000007801 UP000015103 UP000075885 UP000007819
UP000053105 UP000009046 UP000215335 UP000242457 UP000005203 UP000000311 UP000030765 UP000076407 UP000007062 UP000075920 UP000279307 UP000075900 UP000007266 UP000053825 UP000095300 UP000078542 UP000095301 UP000008237 UP000076502 UP000192221 UP000000803 UP000007798 UP000008792 UP000009192 UP000002282 UP000008711 UP000268350 UP000001819 UP000075840 UP000235965 UP000027135 UP000030742 UP000078541 UP000078200 UP000053097 UP000019118 UP000092461 UP000075886 UP000075903 UP000079169 UP000075880 UP000007801 UP000015103 UP000075885 UP000007819
Interpro
Gene 3D
ProteinModelPortal
H9JUF8
A0A2A4IWE8
A0A437BU50
A0A194RED7
A0A212FM53
A0A194PSR1
+ More
A0A0M9ABB5 A0A0C9QVJ3 E0W3N2 A0A232EHU0 A0A1L8DKE8 A0A2A3EI29 A0A088AJT9 A0A310SFA0 U5EP90 E2ASE5 A0A1Q3FYX2 A0A084W753 A0A182XAJ9 Q7QCI6 A0A023ERV0 A0A182W0Q8 A0A3L8D7K5 A0A2M4BPE0 A0A2M4CTE8 A0A2M4A975 A0A182RQJ5 D2A4H2 A0A0L7RJX9 A0A1I8P1T5 A0A195CNT7 T1E8Y1 T1PB93 A0A1Q3FYT5 E2BJJ3 A0A2M4CSE6 A0A2M3ZH04 A0A2M4A985 A0A2M4BP87 A0A1B6HQJ5 A0A154PBV3 A0A1B6F300 A0A1W4WBI3 A0A1B6M6C1 Q961E7 B4MT15 B4MAI4 B4L681 A0A0R1EHZ8 A0A0Q5T2Z1 W8AC87 A0A3B0JWG7 Q29JA5 A0A2C9GR37 A0A0K8TL11 A0A0A1XKV0 D3TMY4 A0A2J7QK37 A0A067REZ3 A0A0K8VCD3 A0A1B6E198 U4UPH0 A0A151JYP6 A0A1A9VB09 A0A026WDA5 N6T2S8 A0A1B0CKC5 A0A1L8DKH8 A0A1Q3G4F5 A0A182QYM8 A0A182VMZ9 A0A1B6K709 A0A0P6JRY3 A0A1S3DUD7 A0A2M4CTQ9 A0A1Y1MNH7 A0A1Q3G4G1 A0A182J7P2 A0A1I8NAV9 A0A1I8P1Z7 A0A2J7QK44 A0A0R1EFD0 A0A0Q5THS5 A0A1W4VZV3 A0A0P9BUP0 T1I9G0 A0A182P3U1 A0A0Q9WES2 X2JEJ4 A0A0Q9XER1 A0A0A1WK05 A0A0P4VKY8 A0A0R3NY12 A0A2S2QMI6 A0A2H8TZN2 E9J9F9 A0A0A9VSD2 J9K8F1 A0A0A9Z894 A0A146L888
A0A0M9ABB5 A0A0C9QVJ3 E0W3N2 A0A232EHU0 A0A1L8DKE8 A0A2A3EI29 A0A088AJT9 A0A310SFA0 U5EP90 E2ASE5 A0A1Q3FYX2 A0A084W753 A0A182XAJ9 Q7QCI6 A0A023ERV0 A0A182W0Q8 A0A3L8D7K5 A0A2M4BPE0 A0A2M4CTE8 A0A2M4A975 A0A182RQJ5 D2A4H2 A0A0L7RJX9 A0A1I8P1T5 A0A195CNT7 T1E8Y1 T1PB93 A0A1Q3FYT5 E2BJJ3 A0A2M4CSE6 A0A2M3ZH04 A0A2M4A985 A0A2M4BP87 A0A1B6HQJ5 A0A154PBV3 A0A1B6F300 A0A1W4WBI3 A0A1B6M6C1 Q961E7 B4MT15 B4MAI4 B4L681 A0A0R1EHZ8 A0A0Q5T2Z1 W8AC87 A0A3B0JWG7 Q29JA5 A0A2C9GR37 A0A0K8TL11 A0A0A1XKV0 D3TMY4 A0A2J7QK37 A0A067REZ3 A0A0K8VCD3 A0A1B6E198 U4UPH0 A0A151JYP6 A0A1A9VB09 A0A026WDA5 N6T2S8 A0A1B0CKC5 A0A1L8DKH8 A0A1Q3G4F5 A0A182QYM8 A0A182VMZ9 A0A1B6K709 A0A0P6JRY3 A0A1S3DUD7 A0A2M4CTQ9 A0A1Y1MNH7 A0A1Q3G4G1 A0A182J7P2 A0A1I8NAV9 A0A1I8P1Z7 A0A2J7QK44 A0A0R1EFD0 A0A0Q5THS5 A0A1W4VZV3 A0A0P9BUP0 T1I9G0 A0A182P3U1 A0A0Q9WES2 X2JEJ4 A0A0Q9XER1 A0A0A1WK05 A0A0P4VKY8 A0A0R3NY12 A0A2S2QMI6 A0A2H8TZN2 E9J9F9 A0A0A9VSD2 J9K8F1 A0A0A9Z894 A0A146L888
PDB
2Y7J
E-value=2.90216e-103,
Score=959
Ontologies
GO
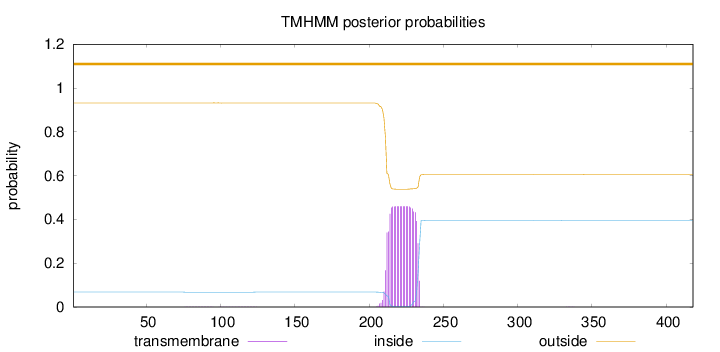
Topology
Length:
418
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.99702999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06805
outside
1 - 418
Population Genetic Test Statistics
Pi
189.588847
Theta
18.200465
Tajima's D
0.749567
CLR
0.448854
CSRT
0.596220188990551
Interpretation
Uncertain
