Gene
KWMTBOMO09921
Annotation
S33901_reverse_transcriptase_-_silkworm_transposon_Pao
Location in the cell
Mitochondrial Reliability : 1.932 Nuclear Reliability : 2.219
Sequence
CDS
ATGAACTCCTTTACAAAAACAGAAAATATAAACAGACTCAGAAGGAACCTGAAAGGAAGGGCAAAGGAAGCCGTTGACGGATTACTTATAACAAACGCTGATCCGTCCGACGTCATAAGAAGTCTAGAAGCGCGATTCGGAAGACCGGAAACGATAGCCATAACGGAGTTAGACACGCTACGAGCTCTGCCAAGACTAACAGAAACACCAAGAGACATTTGTATATTCTCCAGTAAGGTGACCACCGCCGTAGCTACGCTTCGTGCATTAAATTGCACACATTACTTATATAATCCGGAAACTACCAAAACAATGTTAGAAAAACTCACACCAACACTACGTTACCGATACTACGACTTCACCGCGATACAACCGAAGGAGGATCCGGATCTGATTAAATTTGAAAAATTCATGAAAAGAGAAGCCGAACTGTGCAGCCCTTATGCACAGCCTGAACAGGCGGGGCACTACTCGCAGCCCGCACAACATAACAGACGCACACAAAACGTGCACATAGTCAGTGAGAAGCCATCACGAGCTAAATGTCCGGTATGTAGCAACACCGAACACACCACAACAGACTGCTACATATTCAAGAAGGCAGACTCAAACACAAGATGGGACATCGCTAAGAATAAACACCTGTGTTTCCGATGTCTCAAGTATAGAAATAAAACCCACAACTGTAGACCGAAGACTTGTGGCATCAATGATTGCAAATATACTCACAACAAGATGCTACACTTCGACAGAAAAATTGAAAAAACAGACAACAGTGACAAGGAAACAACAGAGAACATTAATTCCGCTTGGACCGGAAAACAGAAACAGTCCTATTTGAAAATAATCCCAGTCCAAGTACAAGGACCGATAGGCACGGTTGATACATACGCGCTGCTCGACGATGGATCAACGGTAACACTGATAGACGAAATTATTTGCAAGAAGACTGGAATAACAGGACCAATCGATCCGTTACACATACAGGCGATTAACAACATAAAATCAACGGAAACAAGGTCAAGAAGAGTCAACCTCACGCTCAGAGGCCTCAACAGTCGAAAAGAAATAATACAAGCGAGAACAGTTAACGACCTACAAGTAACAGCACAAAAAATACCAAAGGAACAGATAGATGAGTATTCGCACCTACGAGACATCAGTGACATCATCACGTACGAGAACGCGAAACCTGGAATCCTGATTGGCCAAGACAACTGGCACATGTTACTAACTTCAAAAGTTAGACGAGGCAACAGGAATCAGCCAATAGCGTCACTGACCCCTCTAGGCTGGGTATTGCATGGAGGTCGCACTCGTACCCTAGGCCACCACATAAACTACATAAATCATGCTAGCGAAACCCAGGAAGATGATAAAATAGAAAATCTGGTAAAACAGTATTTCGCTATGGATGCGCTATGCATCACACCAAGAAGACCAAAAACAGACCCAGAGGAACAGGCGCTTCGCATCCTCAACAGCAATACAGTCCACACAACAGATGGAAGATACGAAACTGCTCTGCTCTGGAAAACAGATAATGTCAGTCTACCAGACAACTACAATAACTCGTTAAAGCGACTGATAAATATAGAAAACAAACTCGATCGTAATCTGGAACTGAAACAGAAATACACAGAACAGATGGAAGCACTCGTCGCGAAAGGCTACGCCGAGCCCGCTCCAAAAACAAAAACAGAGAACAGAACGTGGTATCTACCTCACTTTGCCGTCGTGAACCCCCTGAAGCCGGAAAAACTCCGAGTTGTCCACGACGCCGCCGCCAGAACAGGAGGGGTAGCTTTGAATTATATGCTGCTTAAGGGACCGGACGTACTCCAATCACTACCAGGAGTGATAATGCGATTTAGACAGCATAATATAGCAGTAACAGCAGACATCAAAGAGATGTTCATACAAGTAAAATTGAGACCTGAAGACAAAGACGCGCTCCGTTATCTCTGGCGCAAAGATCAGCGAGATGACAAGCCTCCAGAAGAAAACAGAATGACCTCGTTGATCTTCGGTGCGTCAAGTTCTCCTTCCACAGCAATATATGTAAAGAACTTGACCGCCCAGAAACAAGAAGCCACGCACCCGGAGGCGGCAGTCGCAGTACAGAACAGACACTACGTAGACGACTACTTGGATAACTTCAAAACTTTAAAAGATGCAGTTCACATAACTAAAGACTTTCGTCGAAAACACGAAAGAAAGCCGACCTCGAAGACCTTCTGGACCGACAGTAAGATAGTGTTGAGATGGACAAGGACGGGATCACGCTCGTACAAACCTTACGTCACCCAACGCCTGACAGCTATAGAAGATAGTTCAACAATAAACGGGAGGCGATGGTTACCCACAAAGCACAACGTAGCCGACGACGTGACCCGAGATGTCCCAATGTCGTATCAGAATGAACATAAATGGCTCAAAGGACGAGAATTCCTACGCCAACGACGAGACTCCTGGCCCACGGAGTCGGCGACAGTAACAACAACAAAATTAATAGATAAAGTAAACATAGCAGCTGCAGCACCGGCGGGAGCCTCATGGCCGAGGCGTCCCGACAACGTGGGGCGCACCGTAGACCTCAAGGGTGGAGTTCTACGGAGACCAGTACGAAAACTACTGATCCTGCCCATCGAAGAAGACCATCCTGCACCGAGAAGAATGCGACTGACTCGCACGGCGGGAGTAATGTGCAGGACGAAATAG
Protein
MNSFTKTENINRLRRNLKGRAKEAVDGLLITNADPSDVIRSLEARFGRPETIAITELDTLRALPRLTETPRDICIFSSKVTTAVATLRALNCTHYLYNPETTKTMLEKLTPTLRYRYYDFTAIQPKEDPDLIKFEKFMKREAELCSPYAQPEQAGHYSQPAQHNRRTQNVHIVSEKPSRAKCPVCSNTEHTTTDCYIFKKADSNTRWDIAKNKHLCFRCLKYRNKTHNCRPKTCGINDCKYTHNKMLHFDRKIEKTDNSDKETTENINSAWTGKQKQSYLKIIPVQVQGPIGTVDTYALLDDGSTVTLIDEIICKKTGITGPIDPLHIQAINNIKSTETRSRRVNLTLRGLNSRKEIIQARTVNDLQVTAQKIPKEQIDEYSHLRDISDIITYENAKPGILIGQDNWHMLLTSKVRRGNRNQPIASLTPLGWVLHGGRTRTLGHHINYINHASETQEDDKIENLVKQYFAMDALCITPRRPKTDPEEQALRILNSNTVHTTDGRYETALLWKTDNVSLPDNYNNSLKRLINIENKLDRNLELKQKYTEQMEALVAKGYAEPAPKTKTENRTWYLPHFAVVNPLKPEKLRVVHDAAARTGGVALNYMLLKGPDVLQSLPGVIMRFRQHNIAVTADIKEMFIQVKLRPEDKDALRYLWRKDQRDDKPPEENRMTSLIFGASSSPSTAIYVKNLTAQKQEATHPEAAVAVQNRHYVDDYLDNFKTLKDAVHITKDFRRKHERKPTSKTFWTDSKIVLRWTRTGSRSYKPYVTQRLTAIEDSSTINGRRWLPTKHNVADDVTRDVPMSYQNEHKWLKGREFLRQRRDSWPTESATVTTTKLIDKVNIAAAAPAGASWPRRPDNVGRTVDLKGGVLRRPVRKLLILPIEEDHPAPRRMRLTRTAGVMCRTK
Summary
Uniprot
V9GZQ1
Q2MGA5
A0A3S2N5J7
A0A2A4IX21
A0A1W7R681
A0A1Y1LL75
+ More
A0A226DIM6 A0A226DH22 A0A182G158 A0A1W7R699 A0A226DM06 A0A226EKF8 A0A2M4A568 A0A2M4A5H2 A0A2M4A5G3 A0A226EDU9 A0A2M4A5C7 A0A0N1IGH3 A0A2M4A5D7 A0A226EEI4 A0A226E2P7 A0A226D351 A0A1W7R6M1 A0A182H5B8 A0A453YZM1 W8AJC8 A0A182HDB0 K7JCQ3 A0A0K3CPX7 A0A2M4AQD0 A0A182HEF9 A0A2M4CKI9 A0A2M3Z5X5 A0A2M4CKZ0 A0A1Y1LL65 A0A2M4B8T5 A0A1Y1LN76 A0A2M4B8I4 A0A2M4CKZ8 A0A1W7R6A0 A0A2M4B931 A0A2M4B962 A0A2M4CVY8 A0A0N8ES95 A0A182HCB4 A0A1W7R6H4 A0A2M4BAL5 A0A2M4AMX4 A0A2M4B963 A0A2M4B9W9 A0A1W7R6F7 A0A2M4B8V2 A0A2M4B9D0 A0A2M4B9E1 A0A182H4A2 A0A2M4B8T6 A0A1W7R6E5 A0A226DV68 A0A226EFR1 A0A1W7R6D5 W5J8X7 A0A226D8R4 A0A0N5DFH0 A0A182HDR0 W4XEJ7 A0A3R7G4M2 A0A419Q7X7 W4XEJ9 G7Y7I6 A0A2B4R8F3 A0A074ZCE8 A0A085NQS3 A0A0N5DP73 A0A0V1C2G4 A0A085N9I5 A0A0V1LPB1 A0A085N2A4 A0A432I899 A0A085MT01
A0A226DIM6 A0A226DH22 A0A182G158 A0A1W7R699 A0A226DM06 A0A226EKF8 A0A2M4A568 A0A2M4A5H2 A0A2M4A5G3 A0A226EDU9 A0A2M4A5C7 A0A0N1IGH3 A0A2M4A5D7 A0A226EEI4 A0A226E2P7 A0A226D351 A0A1W7R6M1 A0A182H5B8 A0A453YZM1 W8AJC8 A0A182HDB0 K7JCQ3 A0A0K3CPX7 A0A2M4AQD0 A0A182HEF9 A0A2M4CKI9 A0A2M3Z5X5 A0A2M4CKZ0 A0A1Y1LL65 A0A2M4B8T5 A0A1Y1LN76 A0A2M4B8I4 A0A2M4CKZ8 A0A1W7R6A0 A0A2M4B931 A0A2M4B962 A0A2M4CVY8 A0A0N8ES95 A0A182HCB4 A0A1W7R6H4 A0A2M4BAL5 A0A2M4AMX4 A0A2M4B963 A0A2M4B9W9 A0A1W7R6F7 A0A2M4B8V2 A0A2M4B9D0 A0A2M4B9E1 A0A182H4A2 A0A2M4B8T6 A0A1W7R6E5 A0A226DV68 A0A226EFR1 A0A1W7R6D5 W5J8X7 A0A226D8R4 A0A0N5DFH0 A0A182HDR0 W4XEJ7 A0A3R7G4M2 A0A419Q7X7 W4XEJ9 G7Y7I6 A0A2B4R8F3 A0A074ZCE8 A0A085NQS3 A0A0N5DP73 A0A0V1C2G4 A0A085N9I5 A0A0V1LPB1 A0A085N2A4 A0A432I899 A0A085MT01
Pubmed
EMBL
L09635
AAA75005.1
AF530470
AAQ09229.1
RSAL01000375
RVE42106.1
+ More
NWSH01005440 PCG64209.1 GEHC01000967 JAV46678.1 GEZM01052588 GEZM01052587 GEZM01052586 JAV74402.1 LNIX01000019 OXA44537.1 OXA44855.1 JXUM01136032 KQ568367 KXJ69026.1 GEHC01000956 JAV46689.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 GGFK01002635 MBW35956.1 GGFK01002641 MBW35962.1 GGFK01002631 MBW35952.1 LNIX01000004 OXA55408.1 GGFK01002609 MBW35930.1 KQ459986 KPJ18861.1 GGFK01002619 MBW35940.1 OXA55241.1 LNIX01000007 OXA52015.1 LNIX01000039 OXA39294.1 GEHC01000872 JAV46773.1 JXUM01111221 KQ565482 KXJ70922.1 AAAB01008847 GAMC01017930 JAB88625.1 JXUM01034199 KQ561015 KXJ80028.1 AAZX01014669 LN877231 CTR11689.1 GGFK01009497 MBW42818.1 JXUM01005879 KQ560203 KXJ83865.1 GGFL01001682 MBW65860.1 GGFM01003133 MBW23884.1 GGFL01001842 MBW66020.1 JAV74399.1 GGFJ01000302 MBW49443.1 JAV74401.1 GGFJ01000218 MBW49359.1 GGFL01001727 MBW65905.1 GEHC01000947 JAV46698.1 GGFJ01000402 MBW49543.1 GGFJ01000401 MBW49542.1 GGFL01005193 MBW69371.1 GDUN01000090 JAN95829.1 JXUM01127084 KQ567147 KXJ69602.1 GEHC01000914 JAV46731.1 GGFJ01000935 MBW50076.1 GGFK01008809 MBW42130.1 GGFJ01000432 MBW49573.1 GGFJ01000487 MBW49628.1 GEHC01000945 JAV46700.1 GGFJ01000333 MBW49474.1 GGFJ01000471 MBW49612.1 GGFJ01000470 MBW49611.1 JXUM01109099 KQ565290 KXJ71123.1 GGFJ01000306 MBW49447.1 GEHC01000955 JAV46690.1 LNIX01000012 OXA48116.1 OXA56382.1 GEHC01000921 JAV46724.1 ADMH02001934 ETN60411.1 LNIX01000027 OXA41945.1 JXUM01130316 KQ567562 KXJ69370.1 AAGJ04115911 NIRI01000132 RJW72731.1 NIRI01000534 RJW70389.1 DF142918 GAA48921.1 LSMT01001301 PFX12555.1 KL596795 KER24838.1 KL367480 KFD71819.1 JYDH01000001 KRY43517.1 KL367527 KFD66131.1 JYDW01000021 KRZ61212.1 KL367570 KFD63600.1 QNYJ01000054 RUA06460.1 KL367674 KFD60347.1
NWSH01005440 PCG64209.1 GEHC01000967 JAV46678.1 GEZM01052588 GEZM01052587 GEZM01052586 JAV74402.1 LNIX01000019 OXA44537.1 OXA44855.1 JXUM01136032 KQ568367 KXJ69026.1 GEHC01000956 JAV46689.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 GGFK01002635 MBW35956.1 GGFK01002641 MBW35962.1 GGFK01002631 MBW35952.1 LNIX01000004 OXA55408.1 GGFK01002609 MBW35930.1 KQ459986 KPJ18861.1 GGFK01002619 MBW35940.1 OXA55241.1 LNIX01000007 OXA52015.1 LNIX01000039 OXA39294.1 GEHC01000872 JAV46773.1 JXUM01111221 KQ565482 KXJ70922.1 AAAB01008847 GAMC01017930 JAB88625.1 JXUM01034199 KQ561015 KXJ80028.1 AAZX01014669 LN877231 CTR11689.1 GGFK01009497 MBW42818.1 JXUM01005879 KQ560203 KXJ83865.1 GGFL01001682 MBW65860.1 GGFM01003133 MBW23884.1 GGFL01001842 MBW66020.1 JAV74399.1 GGFJ01000302 MBW49443.1 JAV74401.1 GGFJ01000218 MBW49359.1 GGFL01001727 MBW65905.1 GEHC01000947 JAV46698.1 GGFJ01000402 MBW49543.1 GGFJ01000401 MBW49542.1 GGFL01005193 MBW69371.1 GDUN01000090 JAN95829.1 JXUM01127084 KQ567147 KXJ69602.1 GEHC01000914 JAV46731.1 GGFJ01000935 MBW50076.1 GGFK01008809 MBW42130.1 GGFJ01000432 MBW49573.1 GGFJ01000487 MBW49628.1 GEHC01000945 JAV46700.1 GGFJ01000333 MBW49474.1 GGFJ01000471 MBW49612.1 GGFJ01000470 MBW49611.1 JXUM01109099 KQ565290 KXJ71123.1 GGFJ01000306 MBW49447.1 GEHC01000955 JAV46690.1 LNIX01000012 OXA48116.1 OXA56382.1 GEHC01000921 JAV46724.1 ADMH02001934 ETN60411.1 LNIX01000027 OXA41945.1 JXUM01130316 KQ567562 KXJ69370.1 AAGJ04115911 NIRI01000132 RJW72731.1 NIRI01000534 RJW70389.1 DF142918 GAA48921.1 LSMT01001301 PFX12555.1 KL596795 KER24838.1 KL367480 KFD71819.1 JYDH01000001 KRY43517.1 KL367527 KFD66131.1 JYDW01000021 KRZ61212.1 KL367570 KFD63600.1 QNYJ01000054 RUA06460.1 KL367674 KFD60347.1
Proteomes
PRIDE
Pfam
Interpro
IPR005312
DUF1759
+ More
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR041588 Integrase_H2C2
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001841 Znf_RING
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR002999 Tudor
IPR000048 IQ_motif_EF-hand-BS
IPR000253 FHA_dom
IPR041373 RT_RNaseH
IPR003961 FN3_dom
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR003591 Leu-rich_rpt_typical-subtyp
IPR023631 Amidase_dom
IPR036928 AS_sf
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR041588 Integrase_H2C2
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001841 Znf_RING
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR002999 Tudor
IPR000048 IQ_motif_EF-hand-BS
IPR000253 FHA_dom
IPR041373 RT_RNaseH
IPR003961 FN3_dom
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR003591 Leu-rich_rpt_typical-subtyp
IPR023631 Amidase_dom
IPR036928 AS_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
V9GZQ1
Q2MGA5
A0A3S2N5J7
A0A2A4IX21
A0A1W7R681
A0A1Y1LL75
+ More
A0A226DIM6 A0A226DH22 A0A182G158 A0A1W7R699 A0A226DM06 A0A226EKF8 A0A2M4A568 A0A2M4A5H2 A0A2M4A5G3 A0A226EDU9 A0A2M4A5C7 A0A0N1IGH3 A0A2M4A5D7 A0A226EEI4 A0A226E2P7 A0A226D351 A0A1W7R6M1 A0A182H5B8 A0A453YZM1 W8AJC8 A0A182HDB0 K7JCQ3 A0A0K3CPX7 A0A2M4AQD0 A0A182HEF9 A0A2M4CKI9 A0A2M3Z5X5 A0A2M4CKZ0 A0A1Y1LL65 A0A2M4B8T5 A0A1Y1LN76 A0A2M4B8I4 A0A2M4CKZ8 A0A1W7R6A0 A0A2M4B931 A0A2M4B962 A0A2M4CVY8 A0A0N8ES95 A0A182HCB4 A0A1W7R6H4 A0A2M4BAL5 A0A2M4AMX4 A0A2M4B963 A0A2M4B9W9 A0A1W7R6F7 A0A2M4B8V2 A0A2M4B9D0 A0A2M4B9E1 A0A182H4A2 A0A2M4B8T6 A0A1W7R6E5 A0A226DV68 A0A226EFR1 A0A1W7R6D5 W5J8X7 A0A226D8R4 A0A0N5DFH0 A0A182HDR0 W4XEJ7 A0A3R7G4M2 A0A419Q7X7 W4XEJ9 G7Y7I6 A0A2B4R8F3 A0A074ZCE8 A0A085NQS3 A0A0N5DP73 A0A0V1C2G4 A0A085N9I5 A0A0V1LPB1 A0A085N2A4 A0A432I899 A0A085MT01
A0A226DIM6 A0A226DH22 A0A182G158 A0A1W7R699 A0A226DM06 A0A226EKF8 A0A2M4A568 A0A2M4A5H2 A0A2M4A5G3 A0A226EDU9 A0A2M4A5C7 A0A0N1IGH3 A0A2M4A5D7 A0A226EEI4 A0A226E2P7 A0A226D351 A0A1W7R6M1 A0A182H5B8 A0A453YZM1 W8AJC8 A0A182HDB0 K7JCQ3 A0A0K3CPX7 A0A2M4AQD0 A0A182HEF9 A0A2M4CKI9 A0A2M3Z5X5 A0A2M4CKZ0 A0A1Y1LL65 A0A2M4B8T5 A0A1Y1LN76 A0A2M4B8I4 A0A2M4CKZ8 A0A1W7R6A0 A0A2M4B931 A0A2M4B962 A0A2M4CVY8 A0A0N8ES95 A0A182HCB4 A0A1W7R6H4 A0A2M4BAL5 A0A2M4AMX4 A0A2M4B963 A0A2M4B9W9 A0A1W7R6F7 A0A2M4B8V2 A0A2M4B9D0 A0A2M4B9E1 A0A182H4A2 A0A2M4B8T6 A0A1W7R6E5 A0A226DV68 A0A226EFR1 A0A1W7R6D5 W5J8X7 A0A226D8R4 A0A0N5DFH0 A0A182HDR0 W4XEJ7 A0A3R7G4M2 A0A419Q7X7 W4XEJ9 G7Y7I6 A0A2B4R8F3 A0A074ZCE8 A0A085NQS3 A0A0N5DP73 A0A0V1C2G4 A0A085N9I5 A0A0V1LPB1 A0A085N2A4 A0A432I899 A0A085MT01
Ontologies
KEGG
GO
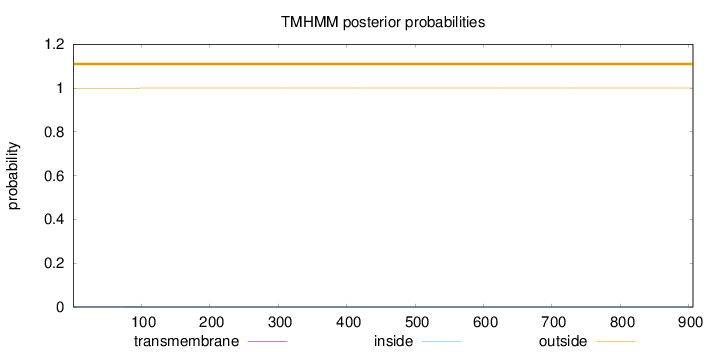
Topology
Length:
906
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01479
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00069
outside
1 - 906
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
13.677437
CSRT
0
Interpretation
Uncertain
