Pre Gene Modal
BGIBMGA013170
Annotation
PREDICTED:_WSCD_family_member_AAEL009094_isoform_X1_[Bombyx_mori]
Full name
WSCD family member AAEL009094
+ More
WSCD family member AGAP003962
WSCD family member GA21586
WSCD family member CG9164
Sulfotransferase
WSCD family member AGAP003962
WSCD family member GA21586
WSCD family member CG9164
Sulfotransferase
Location in the cell
Mitochondrial Reliability : 1.915 PlasmaMembrane Reliability : 1.788
Sequence
CDS
ATGTCTCGTCGTACGATATTCTTGTTCGCGGCGGCTGTCGTCATGCTGTATTTTTCAATAATCTTGTTTATGTTGAGTCCTCTACACGGTACGAAAAGTGCCTATTACGGCAACAACTATATGAATCATTTCGGCTTTCGACAAATACCCAGCGTTAACTGGTGTCATGAGTTGCGTTGGCAGTCGCCGCCTTCACCGCACGTTGTTGCCCTGGCCTCCTACCCTGGAAGCGGCAATACATGGTTCAGATATCTTCTACAGCAGGCCACAGGTATAGTCACGGGCTCTGTGTACATGGATTATGGACTTCGAGTCCATGGGTTCCCAGCTGAGAATGTGACAGATGGTTCAGTGCTGGTAGTTAAAACTCATGAAGCTCCACCTGCTCATTCTGACAAGTTTAAATCTGCAATATTACTGATAAGAAACCCAAGAGATGCGATATTGGCGGATTTTAACAGACTTCACAAGGGTCACATCGGCACTGCTCCTAAGTCGGCATTCAAACGAATATCGCACGACAAAAAAACCGAGTGGTGGTGGCACGTGCGCCGCGAGGTGGTGTCGTGGGAGGCGCTGCACAGGGCCTGGCTGGCGGGGTTCCGCGGCGCGCTGCTGGTGCTGCGCTACGAGCAGCTGGCGGAGGACCCCGGGCCGCCGCTGCAGGCCGCGCTGGCCTTCCTCAACCGCACCGTCCCGCAGGAAAGCTTGGAGTGCGCGCTGGCGAACAAGGAGGGCATTTACCGACGCCGAAACAAGCATCAGAACTTCGATCCCTTCACTCCGCAGATGTACGAGCTGCTGGACGCGGCGCAGGCCAGGGTCGCGGGGCTGGTGGCGCGCCGCACGCGTGACGTCACCACGCGCCCACCCACCCGGGCTTAA
Protein
MSRRTIFLFAAAVVMLYFSIILFMLSPLHGTKSAYYGNNYMNHFGFRQIPSVNWCHELRWQSPPSPHVVALASYPGSGNTWFRYLLQQATGIVTGSVYMDYGLRVHGFPAENVTDGSVLVVKTHEAPPAHSDKFKSAILLIRNPRDAILADFNRLHKGHIGTAPKSAFKRISHDKKTEWWWHVRREVVSWEALHRAWLAGFRGALLVLRYEQLAEDPGPPLQAALAFLNRTVPQESLECALANKEGIYRRRNKHQNFDPFTPQMYELLDAAQARVAGLVARRTRDVTTRPPTRA
Summary
Similarity
Belongs to the WSCD family.
Belongs to the sulfotransferase 1 family.
Belongs to the sulfotransferase 1 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Alternative splicing
Feature
chain WSCD family member AAEL009094
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JUF7
A0A437BUE3
A0A2H1V803
A0A2W1BE10
A0A0N0PBX5
A0A2A4JJ84
+ More
A0A194PKA9 A0A212F232 A0A0N1PFY4 A0A194PIM0 Q16WU7 A0A0K8WGS9 A0A0K8W8M0 A0A0K8UXV4 A0A194PIC8 W5J3V8 A0A182FA94 E0W3N7 Q7Q297 A0A336MS04 D2A4G1 A0A182RWV7 A0A2J7RGN9 A0A182VQH7 A0A182PZX8 A0A1I8MIT8 A0A182N6R6 A0A182M101 A0A084VGD5 A0A182G1G6 A0A1I8P216 A0A182PBY4 B4M7V1 A0A1I8P208 A0A182U5V9 B4L2I6 A0A182I8C4 A0A182XH28 Q7Q297-2 W8C462 A0A182KMU2 A0A182XXM5 A0A1B0CIB8 A0A1J1INX8 A0A067R847 A0A444SGY3 D3TQS4 A0A1B6DAR1 A0A336MU61 A0A0L0BL68 A0A1B0GBB8 A0A182J434 J3JTP4 A0A3B0JDT2 A0A3B0JZY2 B3MR93 A0A0M4ENT3 Q29G54 A0A0R3P1T0 B4GVY4 U4U9Z9 B3NTL8 C0MM62 C0MM61 Q9VXV9 B4PWM2 B4R571 B4ILH0 C0MM65 C0MM59 C0MM60 A0A1B6JQ21 A0A1D2N0P8 A0A0K8VIN8 A0A1S3DDX4 V4AKI4 A0A210QIX4 A0A2D0SSL8 A0A1X7V936 A0A3B1J9P7 A0A2R8PXJ4 E9QD45 E7F082 A0A3L7GNW1 A0A3P9DBB5 A0A3Q2WRG7 A0A3B4GDL6
A0A194PKA9 A0A212F232 A0A0N1PFY4 A0A194PIM0 Q16WU7 A0A0K8WGS9 A0A0K8W8M0 A0A0K8UXV4 A0A194PIC8 W5J3V8 A0A182FA94 E0W3N7 Q7Q297 A0A336MS04 D2A4G1 A0A182RWV7 A0A2J7RGN9 A0A182VQH7 A0A182PZX8 A0A1I8MIT8 A0A182N6R6 A0A182M101 A0A084VGD5 A0A182G1G6 A0A1I8P216 A0A182PBY4 B4M7V1 A0A1I8P208 A0A182U5V9 B4L2I6 A0A182I8C4 A0A182XH28 Q7Q297-2 W8C462 A0A182KMU2 A0A182XXM5 A0A1B0CIB8 A0A1J1INX8 A0A067R847 A0A444SGY3 D3TQS4 A0A1B6DAR1 A0A336MU61 A0A0L0BL68 A0A1B0GBB8 A0A182J434 J3JTP4 A0A3B0JDT2 A0A3B0JZY2 B3MR93 A0A0M4ENT3 Q29G54 A0A0R3P1T0 B4GVY4 U4U9Z9 B3NTL8 C0MM62 C0MM61 Q9VXV9 B4PWM2 B4R571 B4ILH0 C0MM65 C0MM59 C0MM60 A0A1B6JQ21 A0A1D2N0P8 A0A0K8VIN8 A0A1S3DDX4 V4AKI4 A0A210QIX4 A0A2D0SSL8 A0A1X7V936 A0A3B1J9P7 A0A2R8PXJ4 E9QD45 E7F082 A0A3L7GNW1 A0A3P9DBB5 A0A3Q2WRG7 A0A3B4GDL6
EC Number
2.8.2.-
Pubmed
19121390
28756777
26354079
22118469
17510324
20920257
+ More
23761445 20566863 12364791 18362917 19820115 25315136 24438588 26483478 17994087 24495485 20966253 25244985 24845553 20353571 26108605 22516182 18057021 15632085 23537049 19126864 10731132 12537572 12537569 18477586 17550304 27289101 23254933 28812685 25329095 23594743 29704459 25186727
23761445 20566863 12364791 18362917 19820115 25315136 24438588 26483478 17994087 24495485 20966253 25244985 24845553 20353571 26108605 22516182 18057021 15632085 23537049 19126864 10731132 12537572 12537569 18477586 17550304 27289101 23254933 28812685 25329095 23594743 29704459 25186727
EMBL
BABH01022753
RSAL01000008
RVE53976.1
ODYU01001161
SOQ36983.1
KZ150106
+ More
PZC73412.1 KQ460773 KPJ12441.1 NWSH01001290 PCG71836.1 KQ459602 KPI93164.1 AGBW02010783 OWR47792.1 KPJ12439.1 KPI93167.1 CH477554 GDHF01011684 GDHF01002065 JAI40630.1 JAI50249.1 GDHF01028014 GDHF01004945 JAI24300.1 JAI47369.1 GDHF01020888 JAI31426.1 KPI93166.1 ADMH02002183 ETN58003.1 DS235882 EEB20243.1 AAAB01008978 UFQS01002343 UFQT01002343 SSX13821.1 SSX33242.1 KQ971344 EFA05808.2 NEVH01003757 PNF40015.1 AXCN02000066 AXCM01003268 ATLV01012845 KE524818 KFB37029.1 JXUM01038488 KQ561167 KXJ79491.1 CH940653 EDW62868.1 CH933810 EDW06862.1 APCN01002703 GAMC01005729 JAC00827.1 AJWK01013184 CVRI01000057 CRL01941.1 KK852680 KDR18632.1 SAUD01037289 RXG52620.1 EZ423776 ADD20052.1 GEDC01014507 JAS22791.1 UFQT01002352 SSX33265.1 JRES01001702 KNC20787.1 CCAG010022534 BT126601 AEE61565.1 OUUW01000003 SPP78312.1 SPP78311.1 CH902622 EDV34298.1 KPU75063.1 CP012528 ALC49746.1 CH379064 KRT07084.1 CH479193 EDW26829.1 KB632191 ERL89847.1 CH954180 EDV47022.1 FM246434 FM246435 FM246436 FM246438 FM246439 FM246440 FM246441 FM246442 CAR94360.1 CAR94361.1 CAR94362.1 CAR94364.1 CAR94365.1 CAR94366.1 CAR94367.1 CAR94368.1 FM246433 CAR94359.1 AE014298 AY069087 AM999314 AM999315 AM999316 AM999317 AM999318 AM999319 AM999320 AM999321 AM999322 CM000162 EDX01768.1 CM000366 EDX17972.1 CH480871 EDW53833.1 FM246437 CAR94363.1 FM246431 CAR94357.1 FM246432 CAR94358.1 GECU01006713 JAT00994.1 LJIJ01000315 ODM98863.1 GDHF01021019 GDHF01013560 JAI31295.1 JAI38754.1 KB199652 ESP04709.1 NEDP02003450 OWF48649.1 AL929310 RAZU01001831 RLQ54741.1
PZC73412.1 KQ460773 KPJ12441.1 NWSH01001290 PCG71836.1 KQ459602 KPI93164.1 AGBW02010783 OWR47792.1 KPJ12439.1 KPI93167.1 CH477554 GDHF01011684 GDHF01002065 JAI40630.1 JAI50249.1 GDHF01028014 GDHF01004945 JAI24300.1 JAI47369.1 GDHF01020888 JAI31426.1 KPI93166.1 ADMH02002183 ETN58003.1 DS235882 EEB20243.1 AAAB01008978 UFQS01002343 UFQT01002343 SSX13821.1 SSX33242.1 KQ971344 EFA05808.2 NEVH01003757 PNF40015.1 AXCN02000066 AXCM01003268 ATLV01012845 KE524818 KFB37029.1 JXUM01038488 KQ561167 KXJ79491.1 CH940653 EDW62868.1 CH933810 EDW06862.1 APCN01002703 GAMC01005729 JAC00827.1 AJWK01013184 CVRI01000057 CRL01941.1 KK852680 KDR18632.1 SAUD01037289 RXG52620.1 EZ423776 ADD20052.1 GEDC01014507 JAS22791.1 UFQT01002352 SSX33265.1 JRES01001702 KNC20787.1 CCAG010022534 BT126601 AEE61565.1 OUUW01000003 SPP78312.1 SPP78311.1 CH902622 EDV34298.1 KPU75063.1 CP012528 ALC49746.1 CH379064 KRT07084.1 CH479193 EDW26829.1 KB632191 ERL89847.1 CH954180 EDV47022.1 FM246434 FM246435 FM246436 FM246438 FM246439 FM246440 FM246441 FM246442 CAR94360.1 CAR94361.1 CAR94362.1 CAR94364.1 CAR94365.1 CAR94366.1 CAR94367.1 CAR94368.1 FM246433 CAR94359.1 AE014298 AY069087 AM999314 AM999315 AM999316 AM999317 AM999318 AM999319 AM999320 AM999321 AM999322 CM000162 EDX01768.1 CM000366 EDX17972.1 CH480871 EDW53833.1 FM246437 CAR94363.1 FM246431 CAR94357.1 FM246432 CAR94358.1 GECU01006713 JAT00994.1 LJIJ01000315 ODM98863.1 GDHF01021019 GDHF01013560 JAI31295.1 JAI38754.1 KB199652 ESP04709.1 NEDP02003450 OWF48649.1 AL929310 RAZU01001831 RLQ54741.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000008820 UP000000673 UP000069272 UP000009046 UP000007062 UP000007266 UP000075900 UP000235965 UP000075920 UP000075886 UP000095301 UP000075884 UP000075883 UP000030765 UP000069940 UP000249989 UP000095300 UP000075885 UP000008792 UP000075902 UP000009192 UP000075840 UP000076407 UP000075882 UP000076408 UP000092461 UP000183832 UP000027135 UP000288706 UP000037069 UP000092444 UP000075880 UP000268350 UP000007801 UP000092553 UP000001819 UP000008744 UP000030742 UP000008711 UP000000803 UP000002282 UP000000304 UP000001292 UP000094527 UP000079169 UP000030746 UP000242188 UP000221080 UP000007879 UP000018467 UP000000437 UP000273346 UP000265160 UP000264840 UP000261460
UP000008820 UP000000673 UP000069272 UP000009046 UP000007062 UP000007266 UP000075900 UP000235965 UP000075920 UP000075886 UP000095301 UP000075884 UP000075883 UP000030765 UP000069940 UP000249989 UP000095300 UP000075885 UP000008792 UP000075902 UP000009192 UP000075840 UP000076407 UP000075882 UP000076408 UP000092461 UP000183832 UP000027135 UP000288706 UP000037069 UP000092444 UP000075880 UP000268350 UP000007801 UP000092553 UP000001819 UP000008744 UP000030742 UP000008711 UP000000803 UP000002282 UP000000304 UP000001292 UP000094527 UP000079169 UP000030746 UP000242188 UP000221080 UP000007879 UP000018467 UP000000437 UP000273346 UP000265160 UP000264840 UP000261460
Interpro
ProteinModelPortal
H9JUF7
A0A437BUE3
A0A2H1V803
A0A2W1BE10
A0A0N0PBX5
A0A2A4JJ84
+ More
A0A194PKA9 A0A212F232 A0A0N1PFY4 A0A194PIM0 Q16WU7 A0A0K8WGS9 A0A0K8W8M0 A0A0K8UXV4 A0A194PIC8 W5J3V8 A0A182FA94 E0W3N7 Q7Q297 A0A336MS04 D2A4G1 A0A182RWV7 A0A2J7RGN9 A0A182VQH7 A0A182PZX8 A0A1I8MIT8 A0A182N6R6 A0A182M101 A0A084VGD5 A0A182G1G6 A0A1I8P216 A0A182PBY4 B4M7V1 A0A1I8P208 A0A182U5V9 B4L2I6 A0A182I8C4 A0A182XH28 Q7Q297-2 W8C462 A0A182KMU2 A0A182XXM5 A0A1B0CIB8 A0A1J1INX8 A0A067R847 A0A444SGY3 D3TQS4 A0A1B6DAR1 A0A336MU61 A0A0L0BL68 A0A1B0GBB8 A0A182J434 J3JTP4 A0A3B0JDT2 A0A3B0JZY2 B3MR93 A0A0M4ENT3 Q29G54 A0A0R3P1T0 B4GVY4 U4U9Z9 B3NTL8 C0MM62 C0MM61 Q9VXV9 B4PWM2 B4R571 B4ILH0 C0MM65 C0MM59 C0MM60 A0A1B6JQ21 A0A1D2N0P8 A0A0K8VIN8 A0A1S3DDX4 V4AKI4 A0A210QIX4 A0A2D0SSL8 A0A1X7V936 A0A3B1J9P7 A0A2R8PXJ4 E9QD45 E7F082 A0A3L7GNW1 A0A3P9DBB5 A0A3Q2WRG7 A0A3B4GDL6
A0A194PKA9 A0A212F232 A0A0N1PFY4 A0A194PIM0 Q16WU7 A0A0K8WGS9 A0A0K8W8M0 A0A0K8UXV4 A0A194PIC8 W5J3V8 A0A182FA94 E0W3N7 Q7Q297 A0A336MS04 D2A4G1 A0A182RWV7 A0A2J7RGN9 A0A182VQH7 A0A182PZX8 A0A1I8MIT8 A0A182N6R6 A0A182M101 A0A084VGD5 A0A182G1G6 A0A1I8P216 A0A182PBY4 B4M7V1 A0A1I8P208 A0A182U5V9 B4L2I6 A0A182I8C4 A0A182XH28 Q7Q297-2 W8C462 A0A182KMU2 A0A182XXM5 A0A1B0CIB8 A0A1J1INX8 A0A067R847 A0A444SGY3 D3TQS4 A0A1B6DAR1 A0A336MU61 A0A0L0BL68 A0A1B0GBB8 A0A182J434 J3JTP4 A0A3B0JDT2 A0A3B0JZY2 B3MR93 A0A0M4ENT3 Q29G54 A0A0R3P1T0 B4GVY4 U4U9Z9 B3NTL8 C0MM62 C0MM61 Q9VXV9 B4PWM2 B4R571 B4ILH0 C0MM65 C0MM59 C0MM60 A0A1B6JQ21 A0A1D2N0P8 A0A0K8VIN8 A0A1S3DDX4 V4AKI4 A0A210QIX4 A0A2D0SSL8 A0A1X7V936 A0A3B1J9P7 A0A2R8PXJ4 E9QD45 E7F082 A0A3L7GNW1 A0A3P9DBB5 A0A3Q2WRG7 A0A3B4GDL6
PDB
3NIB
E-value=0.000215629,
Score=104
Ontologies
GO
PANTHER
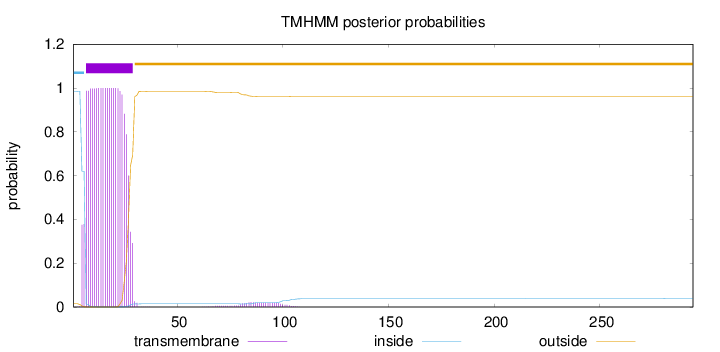
Topology
Subcellular location
Membrane
Length:
294
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.1329199999999
Exp number, first 60 AAs:
21.63144
Total prob of N-in:
0.98559
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 294
Population Genetic Test Statistics
Pi
164.457221
Theta
196.661237
Tajima's D
-0.855147
CLR
1.691271
CSRT
0.171541422928854
Interpretation
Uncertain
