Gene
KWMTBOMO09916
Pre Gene Modal
BGIBMGA013252
Annotation
PREDICTED:_CCR4-NOT_transcription_complex_subunit_10-like_[Amyelois_transitella]
Full name
CCR4-NOT transcription complex subunit 10
Location in the cell
Golgi Reliability : 1.05
Sequence
CDS
ATGTCAAACAACAAAGATCGCGATGAACCTGCTTTTTTAGCTCACCAATGTTTCTTAAAAAAGGACTACACAGCGGCATTACAACATCTTAGCGACTTGGAAAACCTTGTGGGAACTTCGAATAAAAGGGTTCAACACAACAAAGCCGTTGTAGAGTTTATGGCTGGAGAGATGAAGAATGTGGAAAAATTCAAAAATGCCATTACACAGTTATCAGGGTTAAATTATTTGGACGTCGAAGTCAAGGATATGACTTCACCCTGTTTGTTGTATAATTACGCGGTTATTCTGTTCCATTCACGTTATTATTATCAATGTGTTGTCATATTAGAGAAATTATTAAGTTCTAAGAGCATAAAAGATGCGAGATTACTACAGCAAATTATCTTACTTATGTTAGAAGCAACAATATGTCGACGTACTTATGATAAGACTATAGAAATCGCCAAGAACCACGGCGAGCCATTAAAAATTAATAATGAAGTTAGTGAGTTATTTGAAAGGATTATATCACGTGCTCAATTATTAGTTGGCCAAAAAGTAAAATTAAACCTTAAGCCTGATTCCATAGAGAACATATTTGTTATAGCACAGCAACATTACATCAATGGTAATGTAAAGGAAGCAGCTAATATATTGGGATATTACAAAACCTTAAAGTACAATTATGATATGAAAACTCAAGGTGAAGAAATATGGTCTGCCGTAAATAATAATTTGGGTGTCATATATTTATCAATTAAAAAACCTTATTTAGCCTCAAAATATTTTCAGCATGCCATAAAAGAACATTTGAAAGCTATGGAGTGTGAGGACAGTGAAAAATTAATATCTTGTAAAGATAGATCACTTTATGTGTACAATTTGGGGTTAGCACTTTTAGGTACAAATAATTCAGAAGGTGCTTTTGAGTGCCTAGTTGAAGCTGCTAGACATTTTCCTAATAACCCACGAATTTGGCTTCATTTAGCTGAATGCTGTGTAAAGAAATGTTGCAGTGAAGAAGCTCAGCAATACACAGTAAAGAAATTGGGCAGTGGTCCCCACACACGAGTCTTACTCTCAAAAGAAATAAAAGCAAGATATTCCACATCTGGGGAATCTTTTGCAATACCATCATTATCACTGGAATTTGCTGCACTATGTCTACGGAATGCTATGACTCTACTTCCCAACCAAGAACCTCCAGCGGATGTAACTGCATCACTTATACAAGTACCACCAGGTATCCCAATAACCTGGAAACAACGTAGCGAACTCAAGAACTCTGCCCTCGTTTTACAAAGTTATGTATTACTGCACTTGCAAGATCCATTAGCGGCTCTTGTCAGTGCCAATGAGTTACTAGCTCAACCAGACGCATCTAGTAGTCATAAAGCATGGGCTCACATATATGCAGCTGAAGCTCTCATAAATCTTGATCGAATAGCTGATGCTGTGGAGCATCTACATCCACCAACAATTCATGACTTAGTTTCAGTACTGCCGTATCAAATGAGGGATATGATTGCTGTATCTGTGTGGGCTAAAGCTGCAGTGTGTCATATTTTAAGAGGGGATCTAGTAACAGCAAAAAAGATACTTTTACAAATAAATTCACCAAAAGTTCTACCTCTACAGATGTATTTAGAGATTTGCACAGGTAATATAGACAATTGTCACACAATATTGAGAAAGATTAGATACACAAATCCCTCCATTCAATAA
Protein
MSNNKDRDEPAFLAHQCFLKKDYTAALQHLSDLENLVGTSNKRVQHNKAVVEFMAGEMKNVEKFKNAITQLSGLNYLDVEVKDMTSPCLLYNYAVILFHSRYYYQCVVILEKLLSSKSIKDARLLQQIILLMLEATICRRTYDKTIEIAKNHGEPLKINNEVSELFERIISRAQLLVGQKVKLNLKPDSIENIFVIAQQHYINGNVKEAANILGYYKTLKYNYDMKTQGEEIWSAVNNNLGVIYLSIKKPYLASKYFQHAIKEHLKAMECEDSEKLISCKDRSLYVYNLGLALLGTNNSEGAFECLVEAARHFPNNPRIWLHLAECCVKKCCSEEAQQYTVKKLGSGPHTRVLLSKEIKARYSTSGESFAIPSLSLEFAALCLRNAMTLLPNQEPPADVTASLIQVPPGIPITWKQRSELKNSALVLQSYVLLHLQDPLAALVSANELLAQPDASSSHKAWAHIYAAEALINLDRIADAVEHLHPPTIHDLVSVLPYQMRDMIAVSVWAKAAVCHILRGDLVTAKKILLQINSPKVLPLQMYLEICTGNIDNCHTILRKIRYTNPSIQ
Summary
Description
Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Is not required for association of CNOT7 to the CCR4-NOT complex (By similarity).
Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Is not required for association of CNOT7 to the CCR4-NOT complex.
Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Is not required for association of CNOT7 to the CCR4-NOT complex.
Subunit
Component of the CCR4-NOT complex; distinct complexes seem to exist that differ in the participation of probably mutually exclusive catalytic subunits. CNOT10 and CNOT11 form a subcomplex docked to the CNOT1 scaffold (By similarity).
Component of the CCR4-NOT complex; distinct complexes seem to exist that differ in the participation of probably mutually exclusive catalytic subunits. CNOT10 and CNOT11 form a subcomplex docked to the CNOT1 scaffold.
Component of the CCR4-NOT complex; distinct complexes seem to exist that differ in the participation of probably mutually exclusive catalytic subunits. CNOT10 and CNOT11 form a subcomplex docked to the CNOT1 scaffold.
Similarity
Belongs to the CNOT10 family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Nucleus
Reference proteome
RNA-mediated gene silencing
Transcription
Transcription regulation
Translation regulation
Alternative splicing
Coiled coil
Polymorphism
Feature
chain CCR4-NOT transcription complex subunit 10
splice variant In isoform 6.
sequence variant In dbSNP:rs11558687.
splice variant In isoform 6.
sequence variant In dbSNP:rs11558687.
Uniprot
H9JUN9
L0N7B9
A0A2W1BP10
A0A2A4JIB1
A0A2H1V803
A0A0N1I8G4
+ More
A0A194PJF6 A0A0L7KZ21 S4PG32 A0A1E1VXF0 A0A212F249 A0A1Y1M9J2 A0A1Y1MCD7 A0A084VH53 D6WQ07 A0A182PDC0 A0A182YFV7 A0A182IP89 A0A182M8F1 A0A336LTU4 A0A336K921 A0A182TR40 A0A182XN46 A0A182LM74 A0A182HI88 A0A1B6E480 A0A182JYA8 A0A2K5PS57 Q5XIA4 A0A182VH21 A0A1S3HDH2 A0A1S4H2M9 A0A182NBI0 A0A061I5Q3 Q8BH15-3 A0A182VTY6 V4AXB5 Q8BH15-2 A0A1U8C9Q5 A0A1U7QUP2 Q8BH15 M1EIM6 A0A182R3X1 N6UEA0 A0A2I3TE87 U6CUP7 A0A250Y4Q9 A0A154PRE8 A0A061I7V6 A0A2K6UVT1 A0A287AXZ3 A0A2Y9KVR8 L5L3W9 A0A452I509 G3H2P5 Q4R350 A0A2Y9L0Y9 A0A452I544 A0A2Y9KT91 A0A3Q7TQW6 A0A1S3G7E2 A0A3Q7UG43 A0A3Q2HI92 A0A3Q2HMX3 A0A3Q7U2T5 A0A3Q2LID5 A0A0P6JJ98 A0A2I3GAF2 A0A2R8ZSC2 G1QWN2 A0A2R8ZY54 A0A2K6R7D0 A0A2K6DBN9 G3RPD6 A0A2K5YA06 A0A2K5M7K2 G7NYB5 K7BLC0 I0FLB2 Q9H9A5 A0A096NQB5 K7BCN0 A0A2I2ZHA5 A0A2K6R7F2 H2PB90 A0A2I3SDV8 A0A1S3WCH2 A0A2J8WYS3 A0A2R8ZSB5 D2HB05 A0A287A7Y4 A0A2I3S5M4 A0A2I3GMJ1 A0A2K5PSA9 Q7Q1Q8 A0A2Y9G2V1 I3MPA8 Q9H9A5-6 A0A2K5PS20 A0A1S3WD30
A0A194PJF6 A0A0L7KZ21 S4PG32 A0A1E1VXF0 A0A212F249 A0A1Y1M9J2 A0A1Y1MCD7 A0A084VH53 D6WQ07 A0A182PDC0 A0A182YFV7 A0A182IP89 A0A182M8F1 A0A336LTU4 A0A336K921 A0A182TR40 A0A182XN46 A0A182LM74 A0A182HI88 A0A1B6E480 A0A182JYA8 A0A2K5PS57 Q5XIA4 A0A182VH21 A0A1S3HDH2 A0A1S4H2M9 A0A182NBI0 A0A061I5Q3 Q8BH15-3 A0A182VTY6 V4AXB5 Q8BH15-2 A0A1U8C9Q5 A0A1U7QUP2 Q8BH15 M1EIM6 A0A182R3X1 N6UEA0 A0A2I3TE87 U6CUP7 A0A250Y4Q9 A0A154PRE8 A0A061I7V6 A0A2K6UVT1 A0A287AXZ3 A0A2Y9KVR8 L5L3W9 A0A452I509 G3H2P5 Q4R350 A0A2Y9L0Y9 A0A452I544 A0A2Y9KT91 A0A3Q7TQW6 A0A1S3G7E2 A0A3Q7UG43 A0A3Q2HI92 A0A3Q2HMX3 A0A3Q7U2T5 A0A3Q2LID5 A0A0P6JJ98 A0A2I3GAF2 A0A2R8ZSC2 G1QWN2 A0A2R8ZY54 A0A2K6R7D0 A0A2K6DBN9 G3RPD6 A0A2K5YA06 A0A2K5M7K2 G7NYB5 K7BLC0 I0FLB2 Q9H9A5 A0A096NQB5 K7BCN0 A0A2I2ZHA5 A0A2K6R7F2 H2PB90 A0A2I3SDV8 A0A1S3WCH2 A0A2J8WYS3 A0A2R8ZSB5 D2HB05 A0A287A7Y4 A0A2I3S5M4 A0A2I3GMJ1 A0A2K5PSA9 Q7Q1Q8 A0A2Y9G2V1 I3MPA8 Q9H9A5-6 A0A2K5PS20 A0A1S3WD30
Pubmed
19121390
28756777
26354079
26227816
23622113
22118469
+ More
28004739 24438588 18362917 19820115 25244985 20966253 15489334 12364791 23929341 16141072 21183079 23254933 23236062 23537049 16136131 28087693 23258410 28562605 21804562 19892987 22722832 25362486 22398555 22002653 25319552 14702039 16641997 17974005 19413330 19558367 21269460 23232451 23221646 23303381 24275569 20010809
28004739 24438588 18362917 19820115 25244985 20966253 15489334 12364791 23929341 16141072 21183079 23254933 23236062 23537049 16136131 28087693 23258410 28562605 21804562 19892987 22722832 25362486 22398555 22002653 25319552 14702039 16641997 17974005 19413330 19558367 21269460 23232451 23221646 23303381 24275569 20010809
EMBL
BABH01022754
AK289348
BAM73875.1
KZ150106
PZC73413.1
NWSH01001290
+ More
PCG71835.1 ODYU01001161 SOQ36983.1 KQ460773 KPJ12436.1 KQ459602 KPI93168.1 JTDY01004241 KOB68400.1 GAIX01003797 JAA88763.1 GDQN01011648 JAT79406.1 AGBW02010783 OWR47793.1 GEZM01040032 JAV80985.1 GEZM01040030 JAV80987.1 ATLV01013143 KE524841 KFB37297.1 KQ971354 EFA06148.2 AXCM01002025 UFQS01000188 UFQT01000188 SSX00990.1 SSX21370.1 SSX00991.1 SSX21371.1 APCN01005656 GEDC01004558 JAS32740.1 BC083782 AAAB01008980 KE675745 ERE74796.1 AK029103 AK149944 AK161842 BC025554 BC036179 BC039183 BC047204 KB201262 ESO98211.1 JP007862 AER96459.1 APGK01025891 APGK01025892 KB740605 KB632095 ENN80040.1 ERL88756.1 AACZ04000207 AACZ04000208 HAAF01002193 CCP74019.1 GFFV01001328 JAV38617.1 KQ435022 KZC13924.1 ERE74797.1 AEMK02000085 KB030368 ELK17951.1 JH000118 EGW04710.1 AB179418 GEBF01002096 JAO01537.1 ADFV01172028 ADFV01172029 ADFV01172030 ADFV01172031 ADFV01172032 ADFV01172033 ADFV01172034 ADFV01172035 ADFV01172036 ADFV01172037 AJFE02091747 AJFE02091748 AJFE02091749 AJFE02091750 AJFE02091751 AJFE02091752 AJFE02091753 AJFE02091754 AJFE02091755 AJFE02091756 CABD030020604 CABD030020605 CABD030020606 CABD030020607 CABD030020608 CABD030020609 CABD030020610 CABD030020611 CABD030020612 CABD030020613 AQIA01037521 AQIA01037522 AQIA01037523 AQIA01037524 AQIA01037525 AQIA01037526 CM001277 EHH51738.1 GABF01001152 GABD01009970 JAA20993.1 JAA23130.1 JU320281 JU471837 JV045167 CM001254 AFE64037.1 AFH28641.1 AFI35238.1 EHH16818.1 AK021695 AK022576 AK022952 AK023227 AK027026 AK302196 AC138972 AC139452 CH471055 BC002928 BC002931 AL117639 AHZZ02021205 AHZZ02021206 GABC01006926 GABE01000157 NBAG03000225 JAA04412.1 JAA44582.1 PNI73793.1 ABGA01226343 ABGA01226344 ABGA01226345 ABGA01226346 ABGA01226347 ABGA01226348 ABGA01226349 ABGA01226350 ABGA01226351 ABGA01226352 NDHI03003375 PNJ74916.1 GL192643 EFB27836.1 PNI73796.1 EAA14533.4 AGTP01006691 AGTP01006692 AGTP01006693 AGTP01006694 AGTP01006695 AGTP01006696
PCG71835.1 ODYU01001161 SOQ36983.1 KQ460773 KPJ12436.1 KQ459602 KPI93168.1 JTDY01004241 KOB68400.1 GAIX01003797 JAA88763.1 GDQN01011648 JAT79406.1 AGBW02010783 OWR47793.1 GEZM01040032 JAV80985.1 GEZM01040030 JAV80987.1 ATLV01013143 KE524841 KFB37297.1 KQ971354 EFA06148.2 AXCM01002025 UFQS01000188 UFQT01000188 SSX00990.1 SSX21370.1 SSX00991.1 SSX21371.1 APCN01005656 GEDC01004558 JAS32740.1 BC083782 AAAB01008980 KE675745 ERE74796.1 AK029103 AK149944 AK161842 BC025554 BC036179 BC039183 BC047204 KB201262 ESO98211.1 JP007862 AER96459.1 APGK01025891 APGK01025892 KB740605 KB632095 ENN80040.1 ERL88756.1 AACZ04000207 AACZ04000208 HAAF01002193 CCP74019.1 GFFV01001328 JAV38617.1 KQ435022 KZC13924.1 ERE74797.1 AEMK02000085 KB030368 ELK17951.1 JH000118 EGW04710.1 AB179418 GEBF01002096 JAO01537.1 ADFV01172028 ADFV01172029 ADFV01172030 ADFV01172031 ADFV01172032 ADFV01172033 ADFV01172034 ADFV01172035 ADFV01172036 ADFV01172037 AJFE02091747 AJFE02091748 AJFE02091749 AJFE02091750 AJFE02091751 AJFE02091752 AJFE02091753 AJFE02091754 AJFE02091755 AJFE02091756 CABD030020604 CABD030020605 CABD030020606 CABD030020607 CABD030020608 CABD030020609 CABD030020610 CABD030020611 CABD030020612 CABD030020613 AQIA01037521 AQIA01037522 AQIA01037523 AQIA01037524 AQIA01037525 AQIA01037526 CM001277 EHH51738.1 GABF01001152 GABD01009970 JAA20993.1 JAA23130.1 JU320281 JU471837 JV045167 CM001254 AFE64037.1 AFH28641.1 AFI35238.1 EHH16818.1 AK021695 AK022576 AK022952 AK023227 AK027026 AK302196 AC138972 AC139452 CH471055 BC002928 BC002931 AL117639 AHZZ02021205 AHZZ02021206 GABC01006926 GABE01000157 NBAG03000225 JAA04412.1 JAA44582.1 PNI73793.1 ABGA01226343 ABGA01226344 ABGA01226345 ABGA01226346 ABGA01226347 ABGA01226348 ABGA01226349 ABGA01226350 ABGA01226351 ABGA01226352 NDHI03003375 PNJ74916.1 GL192643 EFB27836.1 PNI73796.1 EAA14533.4 AGTP01006691 AGTP01006692 AGTP01006693 AGTP01006694 AGTP01006695 AGTP01006696
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000030765 UP000007266 UP000075885 UP000076408 UP000075880 UP000075883 UP000075902 UP000076407 UP000075882 UP000075840 UP000075881 UP000233040 UP000002494 UP000075903 UP000085678 UP000075884 UP000030759 UP000000589 UP000075920 UP000030746 UP000189706 UP000075900 UP000019118 UP000030742 UP000002277 UP000076502 UP000233220 UP000008227 UP000248482 UP000010552 UP000291020 UP000001075 UP000233100 UP000286640 UP000081671 UP000002281 UP000001073 UP000240080 UP000233200 UP000233120 UP000001519 UP000233140 UP000233060 UP000009130 UP000005640 UP000028761 UP000001595 UP000079721 UP000007062 UP000248481 UP000005215
UP000030765 UP000007266 UP000075885 UP000076408 UP000075880 UP000075883 UP000075902 UP000076407 UP000075882 UP000075840 UP000075881 UP000233040 UP000002494 UP000075903 UP000085678 UP000075884 UP000030759 UP000000589 UP000075920 UP000030746 UP000189706 UP000075900 UP000019118 UP000030742 UP000002277 UP000076502 UP000233220 UP000008227 UP000248482 UP000010552 UP000291020 UP000001075 UP000233100 UP000286640 UP000081671 UP000002281 UP000001073 UP000240080 UP000233200 UP000233120 UP000001519 UP000233140 UP000233060 UP000009130 UP000005640 UP000028761 UP000001595 UP000079721 UP000007062 UP000248481 UP000005215
Interpro
Gene 3D
ProteinModelPortal
H9JUN9
L0N7B9
A0A2W1BP10
A0A2A4JIB1
A0A2H1V803
A0A0N1I8G4
+ More
A0A194PJF6 A0A0L7KZ21 S4PG32 A0A1E1VXF0 A0A212F249 A0A1Y1M9J2 A0A1Y1MCD7 A0A084VH53 D6WQ07 A0A182PDC0 A0A182YFV7 A0A182IP89 A0A182M8F1 A0A336LTU4 A0A336K921 A0A182TR40 A0A182XN46 A0A182LM74 A0A182HI88 A0A1B6E480 A0A182JYA8 A0A2K5PS57 Q5XIA4 A0A182VH21 A0A1S3HDH2 A0A1S4H2M9 A0A182NBI0 A0A061I5Q3 Q8BH15-3 A0A182VTY6 V4AXB5 Q8BH15-2 A0A1U8C9Q5 A0A1U7QUP2 Q8BH15 M1EIM6 A0A182R3X1 N6UEA0 A0A2I3TE87 U6CUP7 A0A250Y4Q9 A0A154PRE8 A0A061I7V6 A0A2K6UVT1 A0A287AXZ3 A0A2Y9KVR8 L5L3W9 A0A452I509 G3H2P5 Q4R350 A0A2Y9L0Y9 A0A452I544 A0A2Y9KT91 A0A3Q7TQW6 A0A1S3G7E2 A0A3Q7UG43 A0A3Q2HI92 A0A3Q2HMX3 A0A3Q7U2T5 A0A3Q2LID5 A0A0P6JJ98 A0A2I3GAF2 A0A2R8ZSC2 G1QWN2 A0A2R8ZY54 A0A2K6R7D0 A0A2K6DBN9 G3RPD6 A0A2K5YA06 A0A2K5M7K2 G7NYB5 K7BLC0 I0FLB2 Q9H9A5 A0A096NQB5 K7BCN0 A0A2I2ZHA5 A0A2K6R7F2 H2PB90 A0A2I3SDV8 A0A1S3WCH2 A0A2J8WYS3 A0A2R8ZSB5 D2HB05 A0A287A7Y4 A0A2I3S5M4 A0A2I3GMJ1 A0A2K5PSA9 Q7Q1Q8 A0A2Y9G2V1 I3MPA8 Q9H9A5-6 A0A2K5PS20 A0A1S3WD30
A0A194PJF6 A0A0L7KZ21 S4PG32 A0A1E1VXF0 A0A212F249 A0A1Y1M9J2 A0A1Y1MCD7 A0A084VH53 D6WQ07 A0A182PDC0 A0A182YFV7 A0A182IP89 A0A182M8F1 A0A336LTU4 A0A336K921 A0A182TR40 A0A182XN46 A0A182LM74 A0A182HI88 A0A1B6E480 A0A182JYA8 A0A2K5PS57 Q5XIA4 A0A182VH21 A0A1S3HDH2 A0A1S4H2M9 A0A182NBI0 A0A061I5Q3 Q8BH15-3 A0A182VTY6 V4AXB5 Q8BH15-2 A0A1U8C9Q5 A0A1U7QUP2 Q8BH15 M1EIM6 A0A182R3X1 N6UEA0 A0A2I3TE87 U6CUP7 A0A250Y4Q9 A0A154PRE8 A0A061I7V6 A0A2K6UVT1 A0A287AXZ3 A0A2Y9KVR8 L5L3W9 A0A452I509 G3H2P5 Q4R350 A0A2Y9L0Y9 A0A452I544 A0A2Y9KT91 A0A3Q7TQW6 A0A1S3G7E2 A0A3Q7UG43 A0A3Q2HI92 A0A3Q2HMX3 A0A3Q7U2T5 A0A3Q2LID5 A0A0P6JJ98 A0A2I3GAF2 A0A2R8ZSC2 G1QWN2 A0A2R8ZY54 A0A2K6R7D0 A0A2K6DBN9 G3RPD6 A0A2K5YA06 A0A2K5M7K2 G7NYB5 K7BLC0 I0FLB2 Q9H9A5 A0A096NQB5 K7BCN0 A0A2I2ZHA5 A0A2K6R7F2 H2PB90 A0A2I3SDV8 A0A1S3WCH2 A0A2J8WYS3 A0A2R8ZSB5 D2HB05 A0A287A7Y4 A0A2I3S5M4 A0A2I3GMJ1 A0A2K5PSA9 Q7Q1Q8 A0A2Y9G2V1 I3MPA8 Q9H9A5-6 A0A2K5PS20 A0A1S3WD30
Ontologies
GO
PANTHER
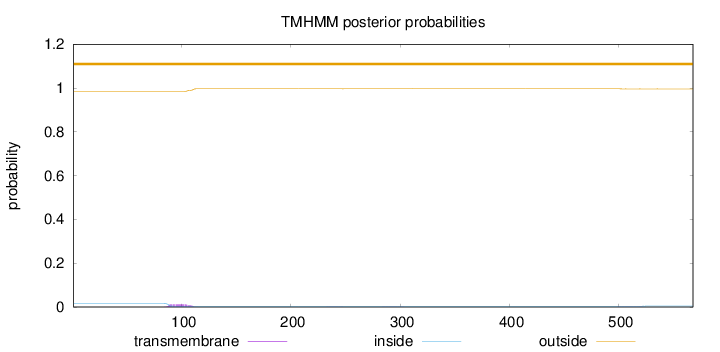
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
568
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.283640000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01606
outside
1 - 568
Population Genetic Test Statistics
Pi
14.149505
Theta
14.293748
Tajima's D
0.219116
CLR
0.588536
CSRT
0.43712814359282
Interpretation
Uncertain
