Gene
KWMTBOMO09914 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013254
Annotation
PREDICTED:_aldo-keto_reductase_AKR2E4-like_isoform_X1_[Amyelois_transitella]
Full name
Aldo-keto reductase AKR2E4
Alternative Name
3-dehydroecdysone reductase
Aldo-keto reductase 2E
Aldo-keto reductase 2E
Location in the cell
Cytoplasmic Reliability : 2.974
Sequence
CDS
ATGGTTGTAGATGTACCCAGTATTAAACTGTCAAATGGGCAAGACATGCCAATTTTAGGGCTCGGAACTTATGCGAGGAAACCAGAGCCTGGTCAAGTTTTGAACTCGGTGAGATGGGCCATCGACGCTGGCTACAGGCACATAGACACCGCGACCGTCTACAAGAACGAGGCTGAAGTTGGTGAGGCCATCAGCGAGAAGATCGACCAGGGTGTCGTCAAGCGGGAACAGCTTTTCGTCACCACCAAATTATGGAACGATAAGCACGCGAGGTCTGATGTCCTGCCGACACTGAAGGAGTCCTTAGCAAAACTGCGTTTGGACTACGTCGATTTGTATCTGGTGCACTGGCCTGTGTCTGTCAACGACAAAGGTGAGGATCTGGCGATCGATCACCTCGACACGTGGAAGGGCATGGAGGAAGTCCTGGCACTGGGCCTCACGAAGGCCATAGGAGTCTCCAACTTCAACGAGGAGCAGCTCGAGAGGTTGCTCAAGGAAGCCACTGTTAAACCAGTCGTCAACCAAGTCGAAATCAACCCAACACTAACTCAGCACAAACTGGTGAGGTTCTGCCAGGAGAGGTCGGTGGTGCCGGTAGCGTATACTCCGCTCGGTCTGGTCTCGGACGCCCGGCCAGAGTTCGCGGGCCTCGACGTCATCAAAACCGACCCGAAGCTGGGCGGACTAGCCGACAAGTACGGAAAGACCAGGGCTCAAGTCGCCCTTAGGTATCTGACCCAACGCAACATTCCCGTTCTACCGAAATCGTTCACGAAGTCCCGGATCGAACAGAACATCGATATATTCGACTTCACGTTGACCGATGACGAGATGGCGCTTGTCGACGGCTACAACATCGACCACCGCTGCGTACCCTCGAAGGCGTTCGCCCATTTGAAGAATTATCCGTTTTAA
Protein
MVVDVPSIKLSNGQDMPILGLGTYARKPEPGQVLNSVRWAIDAGYRHIDTATVYKNEAEVGEAISEKIDQGVVKREQLFVTTKLWNDKHARSDVLPTLKESLAKLRLDYVDLYLVHWPVSVNDKGEDLAIDHLDTWKGMEEVLALGLTKAIGVSNFNEEQLERLLKEATVKPVVNQVEINPTLTQHKLVRFCQERSVVPVAYTPLGLVSDARPEFAGLDVIKTDPKLGGLADKYGKTRAQVALRYLTQRNIPVLPKSFTKSRIEQNIDIFDFTLTDDEMALVDGYNIDHRCVPSKAFAHLKNYPF
Summary
Description
NADP-dependent oxidoreductase with high 3-dehydroecdysone reductase activity. May play a role in the regulation of molting. Has lower activity with phenylglyoxal and isatin (in vitro). Has no activity with NADH as cosubstrate. Has no activity with nitrobenzaldehyde and 3-hydroxybenzaldehyde.
Biophysicochemical Properties
0.0044 mM for 3-dehydroecdysone
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
3D-structure
Complete proteome
Lipid metabolism
NADP
Oxidoreductase
Reference proteome
Steroid metabolism
Feature
chain Aldo-keto reductase AKR2E4
Uniprot
A0A291P0Y9
A0A291P0U2
A0A2A4IYA3
A0A0F6Q2W6
H9JUP1
A0A194PKB3
+ More
U5KBY4 G9F9F4 S4PY90 A0A2H1V815 A0A212F244 A0A2W1BI87 D2A296 A0A2A4JMB2 A0A291P0T7 D6WCX8 A0A1D2MB39 F4WLB5 D6WBX9 A0A1L8D6L9 V5GT06 G9F9G0 A0A3S2LZY0 A0A154PH71 A0A088AGI5 A0A232EMX7 A0A212FML6 A0A0T6B8Q2 B2DBM2 A0A2H1VGR3 A0A151WRD3 A0A151J3M8 H9JTG9 A0A084WAD5 A0A195CRM5 A0A195FYJ2 R4WNM5 I4DP16 A0A182K591 I4DKQ3 A0A443RGQ3 A0A2W1BC51 F4WLB4 A0A2A4JM15 V9IEZ4 A0A0T6AZ90 A0A182QI37 A0A2H1VAA3 A0A2R7WWN4 A0A2R7WXG0 A0A1W4WL13 A0A1I9WLL7 A0A2A3EI70 A0A182RUK3 A0A158NZF8 A0A2H1VAB6 V5GQ24 A0A194Q090 A0A182LYX7 T1DFA3 E9J719 A0A182YBI3 A0A2M3ZFC7 A0A182N3T0 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 V5GXH0 A0A310S4F9 A0A291P0U1 A0A195BMW2 A0A182YBI5 A0A1W4W9Z3 W5JBU8 F5BYH7 U5KC00 Q17DM5 A0A2M4BTH9 A0A2M4AF69 A0A182FVL2 Q17DM4 A0A195CTD6 A0A291P0X3 J3JZ92 A0A2M4BTI2 A0A182JHG0 A0A182WDU3 A0A182QZ12 A0A151IW05 A0A088AGI6 A0A2A4J3U1 A0A291P0W3 A0A067QJF4 A0A2J7QL56 A0A182SQ44 A0A2A3EJL9 E2AB23 A0A437BCA7
U5KBY4 G9F9F4 S4PY90 A0A2H1V815 A0A212F244 A0A2W1BI87 D2A296 A0A2A4JMB2 A0A291P0T7 D6WCX8 A0A1D2MB39 F4WLB5 D6WBX9 A0A1L8D6L9 V5GT06 G9F9G0 A0A3S2LZY0 A0A154PH71 A0A088AGI5 A0A232EMX7 A0A212FML6 A0A0T6B8Q2 B2DBM2 A0A2H1VGR3 A0A151WRD3 A0A151J3M8 H9JTG9 A0A084WAD5 A0A195CRM5 A0A195FYJ2 R4WNM5 I4DP16 A0A182K591 I4DKQ3 A0A443RGQ3 A0A2W1BC51 F4WLB4 A0A2A4JM15 V9IEZ4 A0A0T6AZ90 A0A182QI37 A0A2H1VAA3 A0A2R7WWN4 A0A2R7WXG0 A0A1W4WL13 A0A1I9WLL7 A0A2A3EI70 A0A182RUK3 A0A158NZF8 A0A2H1VAB6 V5GQ24 A0A194Q090 A0A182LYX7 T1DFA3 E9J719 A0A182YBI3 A0A2M3ZFC7 A0A182N3T0 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 V5GXH0 A0A310S4F9 A0A291P0U1 A0A195BMW2 A0A182YBI5 A0A1W4W9Z3 W5JBU8 F5BYH7 U5KC00 Q17DM5 A0A2M4BTH9 A0A2M4AF69 A0A182FVL2 Q17DM4 A0A195CTD6 A0A291P0X3 J3JZ92 A0A2M4BTI2 A0A182JHG0 A0A182WDU3 A0A182QZ12 A0A151IW05 A0A088AGI6 A0A2A4J3U1 A0A291P0W3 A0A067QJF4 A0A2J7QL56 A0A182SQ44 A0A2A3EJL9 E2AB23 A0A437BCA7
EC Number
1.1.1.-
Pubmed
EMBL
MF687608
ATJ44534.1
MF687572
ATJ44498.1
NWSH01005088
PCG64446.1
+ More
KP237872 AKD01725.1 BABH01022758 BABH01022759 BABH01022760 KQ459602 KPI93169.1 KC007350 AGQ45615.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 GAIX01003703 JAA88857.1 ODYU01001161 SOQ36985.1 AGBW02010783 OWR47810.1 KZ150106 PZC73415.1 KQ971338 EFA02158.1 NWSH01001060 PCG72858.1 MF687576 ATJ44502.1 KQ971311 EEZ99072.1 LJIJ01002106 ODM90218.1 GL888207 EGI65078.1 EEZ99009.1 GEYN01000020 JAV02109.1 GALX01003699 JAB64767.1 JN033705 AEW46858.1 RSAL01000093 RVE47941.1 KQ434902 KZC11162.1 NNAY01003257 OXU19723.1 AGBW02007651 OWR54929.1 LJIG01009120 KRT83716.1 AB264706 BAG30781.1 ODYU01002492 SOQ40039.1 KQ982807 KYQ50361.1 KQ980275 KYN17034.1 AB701383 ATLV01022099 KE525328 KFB47179.1 KQ977349 KYN03393.1 KQ981169 KYN45372.1 AK417251 BAN20466.1 AK403246 BAM19656.1 AK401871 BAM18493.1 NCKU01000718 RWS14444.1 KZ150230 PZC72001.1 EGI65077.1 PCG72859.1 JR040676 AEY59237.1 LJIG01022464 KRT80438.1 AXCN02001099 ODYU01001496 SOQ37780.1 KK855773 PTY23851.1 PTY23850.1 KU932401 APA34037.1 KZ288235 PBC31398.1 ADTU01004584 ADTU01004585 SOQ37779.1 GALX01004754 JAB63712.1 KQ459582 KPI98997.1 AXCM01001195 GAMD01003201 JAA98389.1 GL768421 EFZ11357.1 GGFM01006505 MBW27256.1 AAAB01008804 EAA45511.3 APCN01001457 GALX01003438 JAB65028.1 KQ777390 OAD52082.1 MF687570 ATJ44496.1 KQ976433 KYM87328.1 ADMH02001680 ETN61456.1 JF417982 AEB26313.1 KC007349 AGQ45614.1 CH477292 EAT44550.1 EAT44553.1 EAT44554.1 GGFJ01007171 MBW56312.1 GGFK01006115 MBW39436.1 EAT44551.1 EAT44552.1 KYN03394.1 MF687617 ATJ44543.1 BT128573 AEE63530.1 GGFJ01007172 MBW56313.1 KQ980881 KYN11974.1 NWSH01003180 PCG66817.1 MF687607 ATJ44533.1 KK853419 KDR07716.1 NEVH01013254 PNF29307.1 PBC31399.1 GL438234 EFN69299.1 RVE47938.1
KP237872 AKD01725.1 BABH01022758 BABH01022759 BABH01022760 KQ459602 KPI93169.1 KC007350 AGQ45615.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 GAIX01003703 JAA88857.1 ODYU01001161 SOQ36985.1 AGBW02010783 OWR47810.1 KZ150106 PZC73415.1 KQ971338 EFA02158.1 NWSH01001060 PCG72858.1 MF687576 ATJ44502.1 KQ971311 EEZ99072.1 LJIJ01002106 ODM90218.1 GL888207 EGI65078.1 EEZ99009.1 GEYN01000020 JAV02109.1 GALX01003699 JAB64767.1 JN033705 AEW46858.1 RSAL01000093 RVE47941.1 KQ434902 KZC11162.1 NNAY01003257 OXU19723.1 AGBW02007651 OWR54929.1 LJIG01009120 KRT83716.1 AB264706 BAG30781.1 ODYU01002492 SOQ40039.1 KQ982807 KYQ50361.1 KQ980275 KYN17034.1 AB701383 ATLV01022099 KE525328 KFB47179.1 KQ977349 KYN03393.1 KQ981169 KYN45372.1 AK417251 BAN20466.1 AK403246 BAM19656.1 AK401871 BAM18493.1 NCKU01000718 RWS14444.1 KZ150230 PZC72001.1 EGI65077.1 PCG72859.1 JR040676 AEY59237.1 LJIG01022464 KRT80438.1 AXCN02001099 ODYU01001496 SOQ37780.1 KK855773 PTY23851.1 PTY23850.1 KU932401 APA34037.1 KZ288235 PBC31398.1 ADTU01004584 ADTU01004585 SOQ37779.1 GALX01004754 JAB63712.1 KQ459582 KPI98997.1 AXCM01001195 GAMD01003201 JAA98389.1 GL768421 EFZ11357.1 GGFM01006505 MBW27256.1 AAAB01008804 EAA45511.3 APCN01001457 GALX01003438 JAB65028.1 KQ777390 OAD52082.1 MF687570 ATJ44496.1 KQ976433 KYM87328.1 ADMH02001680 ETN61456.1 JF417982 AEB26313.1 KC007349 AGQ45614.1 CH477292 EAT44550.1 EAT44553.1 EAT44554.1 GGFJ01007171 MBW56312.1 GGFK01006115 MBW39436.1 EAT44551.1 EAT44552.1 KYN03394.1 MF687617 ATJ44543.1 BT128573 AEE63530.1 GGFJ01007172 MBW56313.1 KQ980881 KYN11974.1 NWSH01003180 PCG66817.1 MF687607 ATJ44533.1 KK853419 KDR07716.1 NEVH01013254 PNF29307.1 PBC31399.1 GL438234 EFN69299.1 RVE47938.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000283053
UP000007151
UP000007266
+ More
UP000094527 UP000007755 UP000076502 UP000005203 UP000215335 UP000075809 UP000078492 UP000030765 UP000078542 UP000078541 UP000075881 UP000285301 UP000075886 UP000192223 UP000242457 UP000075900 UP000005205 UP000075883 UP000076408 UP000075884 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000078540 UP000000673 UP000008820 UP000069272 UP000075880 UP000075920 UP000027135 UP000235965 UP000075901 UP000000311
UP000094527 UP000007755 UP000076502 UP000005203 UP000215335 UP000075809 UP000078492 UP000030765 UP000078542 UP000078541 UP000075881 UP000285301 UP000075886 UP000192223 UP000242457 UP000075900 UP000005205 UP000075883 UP000076408 UP000075884 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000078540 UP000000673 UP000008820 UP000069272 UP000075880 UP000075920 UP000027135 UP000235965 UP000075901 UP000000311
Pfam
PF00248 Aldo_ket_red
Interpro
SUPFAM
SSF51430
SSF51430
Gene 3D
ProteinModelPortal
A0A291P0Y9
A0A291P0U2
A0A2A4IYA3
A0A0F6Q2W6
H9JUP1
A0A194PKB3
+ More
U5KBY4 G9F9F4 S4PY90 A0A2H1V815 A0A212F244 A0A2W1BI87 D2A296 A0A2A4JMB2 A0A291P0T7 D6WCX8 A0A1D2MB39 F4WLB5 D6WBX9 A0A1L8D6L9 V5GT06 G9F9G0 A0A3S2LZY0 A0A154PH71 A0A088AGI5 A0A232EMX7 A0A212FML6 A0A0T6B8Q2 B2DBM2 A0A2H1VGR3 A0A151WRD3 A0A151J3M8 H9JTG9 A0A084WAD5 A0A195CRM5 A0A195FYJ2 R4WNM5 I4DP16 A0A182K591 I4DKQ3 A0A443RGQ3 A0A2W1BC51 F4WLB4 A0A2A4JM15 V9IEZ4 A0A0T6AZ90 A0A182QI37 A0A2H1VAA3 A0A2R7WWN4 A0A2R7WXG0 A0A1W4WL13 A0A1I9WLL7 A0A2A3EI70 A0A182RUK3 A0A158NZF8 A0A2H1VAB6 V5GQ24 A0A194Q090 A0A182LYX7 T1DFA3 E9J719 A0A182YBI3 A0A2M3ZFC7 A0A182N3T0 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 V5GXH0 A0A310S4F9 A0A291P0U1 A0A195BMW2 A0A182YBI5 A0A1W4W9Z3 W5JBU8 F5BYH7 U5KC00 Q17DM5 A0A2M4BTH9 A0A2M4AF69 A0A182FVL2 Q17DM4 A0A195CTD6 A0A291P0X3 J3JZ92 A0A2M4BTI2 A0A182JHG0 A0A182WDU3 A0A182QZ12 A0A151IW05 A0A088AGI6 A0A2A4J3U1 A0A291P0W3 A0A067QJF4 A0A2J7QL56 A0A182SQ44 A0A2A3EJL9 E2AB23 A0A437BCA7
U5KBY4 G9F9F4 S4PY90 A0A2H1V815 A0A212F244 A0A2W1BI87 D2A296 A0A2A4JMB2 A0A291P0T7 D6WCX8 A0A1D2MB39 F4WLB5 D6WBX9 A0A1L8D6L9 V5GT06 G9F9G0 A0A3S2LZY0 A0A154PH71 A0A088AGI5 A0A232EMX7 A0A212FML6 A0A0T6B8Q2 B2DBM2 A0A2H1VGR3 A0A151WRD3 A0A151J3M8 H9JTG9 A0A084WAD5 A0A195CRM5 A0A195FYJ2 R4WNM5 I4DP16 A0A182K591 I4DKQ3 A0A443RGQ3 A0A2W1BC51 F4WLB4 A0A2A4JM15 V9IEZ4 A0A0T6AZ90 A0A182QI37 A0A2H1VAA3 A0A2R7WWN4 A0A2R7WXG0 A0A1W4WL13 A0A1I9WLL7 A0A2A3EI70 A0A182RUK3 A0A158NZF8 A0A2H1VAB6 V5GQ24 A0A194Q090 A0A182LYX7 T1DFA3 E9J719 A0A182YBI3 A0A2M3ZFC7 A0A182N3T0 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 V5GXH0 A0A310S4F9 A0A291P0U1 A0A195BMW2 A0A182YBI5 A0A1W4W9Z3 W5JBU8 F5BYH7 U5KC00 Q17DM5 A0A2M4BTH9 A0A2M4AF69 A0A182FVL2 Q17DM4 A0A195CTD6 A0A291P0X3 J3JZ92 A0A2M4BTI2 A0A182JHG0 A0A182WDU3 A0A182QZ12 A0A151IW05 A0A088AGI6 A0A2A4J3U1 A0A291P0W3 A0A067QJF4 A0A2J7QL56 A0A182SQ44 A0A2A3EJL9 E2AB23 A0A437BCA7
PDB
3WCZ
E-value=3.58128e-79,
Score=749
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
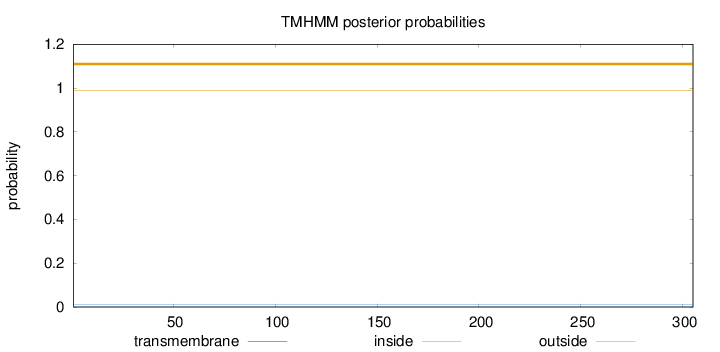
Topology
Length:
305
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01187
outside
1 - 305
Population Genetic Test Statistics
Pi
192.090801
Theta
157.59792
Tajima's D
1.175737
CLR
0.390087
CSRT
0.705664716764162
Interpretation
Uncertain
