Gene
KWMTBOMO09911
Pre Gene Modal
BGIBMGA013255
Annotation
PREDICTED:_SWI/SNF-related_matrix-associated_actin-dependent_regulator_of_chromatin_subfamily_B_member_1-A_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.899
Sequence
CDS
ATGGCCTTACGTACGTACGGCGATAAGCCGATAAGTTTTCAAATCGAAGAAGGAGGAGACTTTTATTGTGTTGGCTCAGAGGTCGGGAATTACCTGCGCTTGTTCCGTGGGTCACTCTACAAGAAGTACCCTGGCATGTGCCGGCGTACCCTCACCAATGAAGAACGTAAGCGATTAATTGACAACGGTCTTGGTCCTCATGTGCTGTCCAGTTCAGTTTCACTGTTGAAGGCGTCAGAAGTTGAAGACATTATTGAAGGGAATGATGATAAATACAAAGCAGTGTCCGTCAGCCAAGAGTTGGCGACACCCCGTGAGAGCAAAAGCAAGAAGCCACACACTCCATCATGGCTGCCAGTCATGCCAAACTCTTCTCATTTGGATGCTGTACCCCAGGCTACTCCAATCAGCAGGAGTAGGGTGCATAATAAGAAGGTGAGAACATTCCCACTGTGTTTCGATGACACAGACATGACTGCAATGCTGGAGAATGCTTCGCAAAAGCAGGTGCTTGTACCGATTCGATTGGACATGGAGATTGAGGGTCAAAAACTCAGGGATACATTTACATGGAATAAAAATGAATCAATAATAACACCAGAACAGTTTGCCGAGGTGCTGTGCGATGATTTAGAGTTGAACACGAGCACCTTCATACCTGCTGTGGCCTCCTCGATCCGGCAACAGATCGATGCCTTCCCGAGCGAGCCTCCCCCCATACTGGAGGAGCATACCGACCAGAGGGTCCTCGTCAAACTCAATATACACGTTGGAAACACTTCGTTGGTAGATCAGGTGGAGTGGGACATGGCCGAGAAGGAGAACAACCCGGAGCAGTTCGCGATGAAGCTGTGCTCGGAGCTGGGCCTCGGCGGGGAGTTCGTGACCGGCATCGCGTACTCCGTGCGCGGGCAGCTCGGCTGGCACCAGCGGACCTACGCCTTCAGCGAGGCGCCCCTACCCGTGGTCGAGACGCCGTACCGCGTCCCGTCGGAGGCCGAGCAGTGGGCCCCGCACCTCGAGACCCTCACCAACGCGGAGATGGAGAAGAAGATCCGGGACCAGGACCGGAACACCAGGCGCATGCGGAGGCTCGCCAACACCGCGCCCGATGTGTCTTTGAGTCGCGCAGTCAACCGGCTGATCAGAGCTGACGAAGCACTGTTCCCGTCCACCCCGGAAAAGCGAAGAAAAAAGATATAA
Protein
MALRTYGDKPISFQIEEGGDFYCVGSEVGNYLRLFRGSLYKKYPGMCRRTLTNEERKRLIDNGLGPHVLSSSVSLLKASEVEDIIEGNDDKYKAVSVSQELATPRESKSKKPHTPSWLPVMPNSSHLDAVPQATPISRSRVHNKKVRTFPLCFDDTDMTAMLENASQKQVLVPIRLDMEIEGQKLRDTFTWNKNESIITPEQFAEVLCDDLELNTSTFIPAVASSIRQQIDAFPSEPPPILEEHTDQRVLVKLNIHVGNTSLVDQVEWDMAEKENNPEQFAMKLCSELGLGGEFVTGIAYSVRGQLGWHQRTYAFSEAPLPVVETPYRVPSEAEQWAPHLETLTNAEMEKKIRDQDRNTRRMRRLANTAPDVSLSRAVNRLIRADEALFPSTPEKRRKKI
Summary
Similarity
Belongs to the SNF5 family.
Uniprot
A0A3S2LN14
A0A194PJG0
A0A2H1V844
S4P996
A0A212F237
H9JUP2
+ More
A0A2W1BJH8 A0A0N1IGL9 A0A026W2P9 A0A151J937 A0A195CBG5 E9J8K5 F4X888 A0A195BG33 A0A0J7L4E0 A0A195FDD4 A0A158NBU7 E2ACH7 A0A151X9S5 E2BIE4 K7IZ56 A0A154P8C2 A0A0M9A1C1 A0A2A3ESG5 V9ILP6 A0A088AH82 A0A1L8DWW0 A0A0C9R9F5 A0A232EUG9 A0A0L7QWF2 A0A2M3ZIF5 A0A2M4AP82 A0A2M4BRJ2 W5J9Z9 A0A182FS92 A0A182QKJ4 A0A1B0DG90 A0A182MRL7 A0A182RFZ1 A0A182YND5 A0A182JT92 A0A182PR88 A0A182VW67 A0A182V9R0 Q7PUT1 A0A182HK49 A0A1W4WLL1 A0A067QTD8 A0A347ZJB3 A0A1W4WWD3 A0A1B6F4F6 A0A1B6JMY0 A0A084W0I9 A0A1B6HS75 A0A1B6LS68 A0A1B6KLN7 J3JTD4 A0A182JE89 A0A182NQY5 D2A622 A0A182TNU2 U4U0D9 A0A2J7QH19 U5ELY9 A0A1Y1K883 A0A1Q3EY24 A0A0T6AWJ3 A0A182GKV8 N6SWY7 A0A0L0BUE9 B0WP88 A0A1S4F7F7 Q17D09 R4QPV6 A0A2R7X529 A0A224XE56 A0A0N7Z9F1 A0A0V0G6H6 A0A023F5T7 A0A2S2NY76 T1I138 A0A069DUV3 W8BU58 A0A1I8NWF5 A0A2S2QZ90 A0A1I8MI70 A0A0K8WF35 A0A034WNW4 J9K0K9 A0A2H8TFS0 A0A1J1ISV8 A0A336KQX1 A0A1S4EHV6 B4JHV6 A0A2P8ZK51 E0VHZ0 B4LZC1 B4KD96 A0A3B0KF71 B5DXY5
A0A2W1BJH8 A0A0N1IGL9 A0A026W2P9 A0A151J937 A0A195CBG5 E9J8K5 F4X888 A0A195BG33 A0A0J7L4E0 A0A195FDD4 A0A158NBU7 E2ACH7 A0A151X9S5 E2BIE4 K7IZ56 A0A154P8C2 A0A0M9A1C1 A0A2A3ESG5 V9ILP6 A0A088AH82 A0A1L8DWW0 A0A0C9R9F5 A0A232EUG9 A0A0L7QWF2 A0A2M3ZIF5 A0A2M4AP82 A0A2M4BRJ2 W5J9Z9 A0A182FS92 A0A182QKJ4 A0A1B0DG90 A0A182MRL7 A0A182RFZ1 A0A182YND5 A0A182JT92 A0A182PR88 A0A182VW67 A0A182V9R0 Q7PUT1 A0A182HK49 A0A1W4WLL1 A0A067QTD8 A0A347ZJB3 A0A1W4WWD3 A0A1B6F4F6 A0A1B6JMY0 A0A084W0I9 A0A1B6HS75 A0A1B6LS68 A0A1B6KLN7 J3JTD4 A0A182JE89 A0A182NQY5 D2A622 A0A182TNU2 U4U0D9 A0A2J7QH19 U5ELY9 A0A1Y1K883 A0A1Q3EY24 A0A0T6AWJ3 A0A182GKV8 N6SWY7 A0A0L0BUE9 B0WP88 A0A1S4F7F7 Q17D09 R4QPV6 A0A2R7X529 A0A224XE56 A0A0N7Z9F1 A0A0V0G6H6 A0A023F5T7 A0A2S2NY76 T1I138 A0A069DUV3 W8BU58 A0A1I8NWF5 A0A2S2QZ90 A0A1I8MI70 A0A0K8WF35 A0A034WNW4 J9K0K9 A0A2H8TFS0 A0A1J1ISV8 A0A336KQX1 A0A1S4EHV6 B4JHV6 A0A2P8ZK51 E0VHZ0 B4LZC1 B4KD96 A0A3B0KF71 B5DXY5
Pubmed
26354079
23622113
22118469
19121390
28756777
24508170
+ More
30249741 21282665 21719571 21347285 20798317 20075255 28648823 20920257 23761445 25244985 12364791 14747013 17210077 24845553 24438588 22516182 18362917 19820115 23537049 28004739 26483478 26108605 17510324 25392228 27129103 25474469 26334808 24495485 25315136 25348373 17994087 29403074 20566863 15632085
30249741 21282665 21719571 21347285 20798317 20075255 28648823 20920257 23761445 25244985 12364791 14747013 17210077 24845553 24438588 22516182 18362917 19820115 23537049 28004739 26483478 26108605 17510324 25392228 27129103 25474469 26334808 24495485 25315136 25348373 17994087 29403074 20566863 15632085
EMBL
RSAL01000051
RVE50214.1
KQ459602
KPI93173.1
ODYU01001161
SOQ36987.1
+ More
GAIX01005601 JAA86959.1 AGBW02010783 OWR47808.1 BABH01022762 KZ150106 PZC73417.1 KQ460773 KPJ12433.1 KK107459 QOIP01000004 EZA50360.1 RLU23612.1 KQ979433 KYN21578.1 KQ978068 KYM97546.1 GL769034 EFZ10853.1 GL888932 EGI57164.1 KQ976500 KYM83164.1 LBMM01000857 KMQ97359.1 KQ981673 KYN38391.1 ADTU01011328 GL438525 EFN68863.1 KQ982373 KYQ57038.1 GL448504 EFN84532.1 KQ434827 KZC07594.1 KQ435794 KOX73883.1 KZ288189 PBC34640.1 JR051830 AEY61567.1 GFDF01003127 JAV10957.1 GBYB01009547 JAG79314.1 NNAY01002138 OXU21957.1 KQ414713 KOC62958.1 GGFM01007504 MBW28255.1 GGFK01009221 MBW42542.1 GGFJ01006566 MBW55707.1 ADMH02002066 ETN59680.1 AXCN02001485 AJVK01033600 AXCM01000964 AAAB01008987 EAA01058.2 APCN01000816 KK853042 KDR12165.1 FX985681 BBA84419.1 GECZ01025016 JAS44753.1 GECU01007274 JAT00433.1 ATLV01019122 KE525262 KFB43733.1 GECU01030211 JAS77495.1 GEBQ01031625 GEBQ01013472 JAT08352.1 JAT26505.1 GEBQ01027853 GEBQ01004753 JAT12124.1 JAT35224.1 BT126490 AEE61454.1 KQ971346 EFA05453.1 KB631749 ERL85763.1 NEVH01014358 PNF27877.1 GANO01001222 JAB58649.1 GEZM01092876 JAV56430.1 GFDL01014842 JAV20203.1 LJIG01022739 KRT78967.1 JXUM01070726 KQ562623 KXJ75470.1 APGK01053516 KB741224 ENN72304.1 JRES01001304 KNC23700.1 DS232020 EDS32133.1 CH477301 EAT44250.1 KC576842 AGL76706.1 KK856874 PTY26520.1 GFTR01005689 JAW10737.1 GDKW01000723 JAI55872.1 GECL01002444 JAP03680.1 GBBI01002174 JAC16538.1 GGMR01009516 MBY22135.1 ACPB03016231 GBGD01001422 JAC87467.1 GAMC01001660 JAC04896.1 GGMS01013790 MBY82993.1 GDHF01002551 JAI49763.1 GAKP01002920 JAC56032.1 ABLF02035782 GFXV01001085 MBW12890.1 CVRI01000057 CRL02190.1 UFQS01000739 UFQT01000739 SSX06492.1 SSX26841.1 CH916369 EDV92864.1 PYGN01000033 PSN56850.1 AAZO01002497 DS235172 EEB12996.1 CH940650 EDW68156.1 CH933806 EDW14878.1 OUUW01000008 SPP84366.1 CM000070 EDY68306.1
GAIX01005601 JAA86959.1 AGBW02010783 OWR47808.1 BABH01022762 KZ150106 PZC73417.1 KQ460773 KPJ12433.1 KK107459 QOIP01000004 EZA50360.1 RLU23612.1 KQ979433 KYN21578.1 KQ978068 KYM97546.1 GL769034 EFZ10853.1 GL888932 EGI57164.1 KQ976500 KYM83164.1 LBMM01000857 KMQ97359.1 KQ981673 KYN38391.1 ADTU01011328 GL438525 EFN68863.1 KQ982373 KYQ57038.1 GL448504 EFN84532.1 KQ434827 KZC07594.1 KQ435794 KOX73883.1 KZ288189 PBC34640.1 JR051830 AEY61567.1 GFDF01003127 JAV10957.1 GBYB01009547 JAG79314.1 NNAY01002138 OXU21957.1 KQ414713 KOC62958.1 GGFM01007504 MBW28255.1 GGFK01009221 MBW42542.1 GGFJ01006566 MBW55707.1 ADMH02002066 ETN59680.1 AXCN02001485 AJVK01033600 AXCM01000964 AAAB01008987 EAA01058.2 APCN01000816 KK853042 KDR12165.1 FX985681 BBA84419.1 GECZ01025016 JAS44753.1 GECU01007274 JAT00433.1 ATLV01019122 KE525262 KFB43733.1 GECU01030211 JAS77495.1 GEBQ01031625 GEBQ01013472 JAT08352.1 JAT26505.1 GEBQ01027853 GEBQ01004753 JAT12124.1 JAT35224.1 BT126490 AEE61454.1 KQ971346 EFA05453.1 KB631749 ERL85763.1 NEVH01014358 PNF27877.1 GANO01001222 JAB58649.1 GEZM01092876 JAV56430.1 GFDL01014842 JAV20203.1 LJIG01022739 KRT78967.1 JXUM01070726 KQ562623 KXJ75470.1 APGK01053516 KB741224 ENN72304.1 JRES01001304 KNC23700.1 DS232020 EDS32133.1 CH477301 EAT44250.1 KC576842 AGL76706.1 KK856874 PTY26520.1 GFTR01005689 JAW10737.1 GDKW01000723 JAI55872.1 GECL01002444 JAP03680.1 GBBI01002174 JAC16538.1 GGMR01009516 MBY22135.1 ACPB03016231 GBGD01001422 JAC87467.1 GAMC01001660 JAC04896.1 GGMS01013790 MBY82993.1 GDHF01002551 JAI49763.1 GAKP01002920 JAC56032.1 ABLF02035782 GFXV01001085 MBW12890.1 CVRI01000057 CRL02190.1 UFQS01000739 UFQT01000739 SSX06492.1 SSX26841.1 CH916369 EDV92864.1 PYGN01000033 PSN56850.1 AAZO01002497 DS235172 EEB12996.1 CH940650 EDW68156.1 CH933806 EDW14878.1 OUUW01000008 SPP84366.1 CM000070 EDY68306.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000005204
UP000053240
UP000053097
+ More
UP000279307 UP000078492 UP000078542 UP000007755 UP000078540 UP000036403 UP000078541 UP000005205 UP000000311 UP000075809 UP000008237 UP000002358 UP000076502 UP000053105 UP000242457 UP000005203 UP000215335 UP000053825 UP000000673 UP000069272 UP000075886 UP000092462 UP000075883 UP000075900 UP000076408 UP000075881 UP000075885 UP000075920 UP000075903 UP000007062 UP000075840 UP000192223 UP000027135 UP000030765 UP000075880 UP000075884 UP000007266 UP000075902 UP000030742 UP000235965 UP000069940 UP000249989 UP000019118 UP000037069 UP000002320 UP000008820 UP000015103 UP000095300 UP000095301 UP000007819 UP000183832 UP000079169 UP000001070 UP000245037 UP000009046 UP000008792 UP000009192 UP000268350 UP000001819
UP000279307 UP000078492 UP000078542 UP000007755 UP000078540 UP000036403 UP000078541 UP000005205 UP000000311 UP000075809 UP000008237 UP000002358 UP000076502 UP000053105 UP000242457 UP000005203 UP000215335 UP000053825 UP000000673 UP000069272 UP000075886 UP000092462 UP000075883 UP000075900 UP000076408 UP000075881 UP000075885 UP000075920 UP000075903 UP000007062 UP000075840 UP000192223 UP000027135 UP000030765 UP000075880 UP000075884 UP000007266 UP000075902 UP000030742 UP000235965 UP000069940 UP000249989 UP000019118 UP000037069 UP000002320 UP000008820 UP000015103 UP000095300 UP000095301 UP000007819 UP000183832 UP000079169 UP000001070 UP000245037 UP000009046 UP000008792 UP000009192 UP000268350 UP000001819
PRIDE
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A3S2LN14
A0A194PJG0
A0A2H1V844
S4P996
A0A212F237
H9JUP2
+ More
A0A2W1BJH8 A0A0N1IGL9 A0A026W2P9 A0A151J937 A0A195CBG5 E9J8K5 F4X888 A0A195BG33 A0A0J7L4E0 A0A195FDD4 A0A158NBU7 E2ACH7 A0A151X9S5 E2BIE4 K7IZ56 A0A154P8C2 A0A0M9A1C1 A0A2A3ESG5 V9ILP6 A0A088AH82 A0A1L8DWW0 A0A0C9R9F5 A0A232EUG9 A0A0L7QWF2 A0A2M3ZIF5 A0A2M4AP82 A0A2M4BRJ2 W5J9Z9 A0A182FS92 A0A182QKJ4 A0A1B0DG90 A0A182MRL7 A0A182RFZ1 A0A182YND5 A0A182JT92 A0A182PR88 A0A182VW67 A0A182V9R0 Q7PUT1 A0A182HK49 A0A1W4WLL1 A0A067QTD8 A0A347ZJB3 A0A1W4WWD3 A0A1B6F4F6 A0A1B6JMY0 A0A084W0I9 A0A1B6HS75 A0A1B6LS68 A0A1B6KLN7 J3JTD4 A0A182JE89 A0A182NQY5 D2A622 A0A182TNU2 U4U0D9 A0A2J7QH19 U5ELY9 A0A1Y1K883 A0A1Q3EY24 A0A0T6AWJ3 A0A182GKV8 N6SWY7 A0A0L0BUE9 B0WP88 A0A1S4F7F7 Q17D09 R4QPV6 A0A2R7X529 A0A224XE56 A0A0N7Z9F1 A0A0V0G6H6 A0A023F5T7 A0A2S2NY76 T1I138 A0A069DUV3 W8BU58 A0A1I8NWF5 A0A2S2QZ90 A0A1I8MI70 A0A0K8WF35 A0A034WNW4 J9K0K9 A0A2H8TFS0 A0A1J1ISV8 A0A336KQX1 A0A1S4EHV6 B4JHV6 A0A2P8ZK51 E0VHZ0 B4LZC1 B4KD96 A0A3B0KF71 B5DXY5
A0A2W1BJH8 A0A0N1IGL9 A0A026W2P9 A0A151J937 A0A195CBG5 E9J8K5 F4X888 A0A195BG33 A0A0J7L4E0 A0A195FDD4 A0A158NBU7 E2ACH7 A0A151X9S5 E2BIE4 K7IZ56 A0A154P8C2 A0A0M9A1C1 A0A2A3ESG5 V9ILP6 A0A088AH82 A0A1L8DWW0 A0A0C9R9F5 A0A232EUG9 A0A0L7QWF2 A0A2M3ZIF5 A0A2M4AP82 A0A2M4BRJ2 W5J9Z9 A0A182FS92 A0A182QKJ4 A0A1B0DG90 A0A182MRL7 A0A182RFZ1 A0A182YND5 A0A182JT92 A0A182PR88 A0A182VW67 A0A182V9R0 Q7PUT1 A0A182HK49 A0A1W4WLL1 A0A067QTD8 A0A347ZJB3 A0A1W4WWD3 A0A1B6F4F6 A0A1B6JMY0 A0A084W0I9 A0A1B6HS75 A0A1B6LS68 A0A1B6KLN7 J3JTD4 A0A182JE89 A0A182NQY5 D2A622 A0A182TNU2 U4U0D9 A0A2J7QH19 U5ELY9 A0A1Y1K883 A0A1Q3EY24 A0A0T6AWJ3 A0A182GKV8 N6SWY7 A0A0L0BUE9 B0WP88 A0A1S4F7F7 Q17D09 R4QPV6 A0A2R7X529 A0A224XE56 A0A0N7Z9F1 A0A0V0G6H6 A0A023F5T7 A0A2S2NY76 T1I138 A0A069DUV3 W8BU58 A0A1I8NWF5 A0A2S2QZ90 A0A1I8MI70 A0A0K8WF35 A0A034WNW4 J9K0K9 A0A2H8TFS0 A0A1J1ISV8 A0A336KQX1 A0A1S4EHV6 B4JHV6 A0A2P8ZK51 E0VHZ0 B4LZC1 B4KD96 A0A3B0KF71 B5DXY5
PDB
6AX5
E-value=8.00924e-27,
Score=299
Ontologies
GO
GO:0005634
GO:0000228
GO:0006338
GO:0035060
GO:0006337
GO:0071565
GO:0071564
GO:0006357
GO:0003713
GO:0016514
GO:0016021
GO:0004386
GO:0097150
GO:0016586
GO:0008586
GO:0045893
GO:0008587
GO:0007474
GO:0070984
GO:0070983
GO:0043697
GO:0007406
GO:0030957
GO:0016020
GO:0015074
GO:0006414
GO:0004816
GO:0005737
GO:0006421
GO:0016155
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
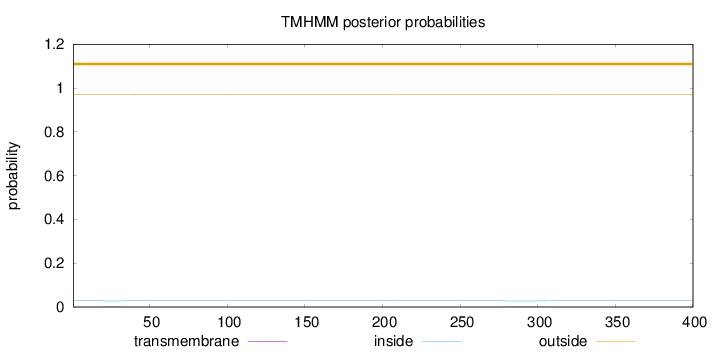
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01679
Exp number, first 60 AAs:
0.00241
Total prob of N-in:
0.02805
outside
1 - 400
Population Genetic Test Statistics
Pi
24.801457
Theta
206.332899
Tajima's D
-1.169162
CLR
1.390555
CSRT
0.110044497775111
Interpretation
Uncertain
