Gene
KWMTBOMO09902
Pre Gene Modal
BGIBMGA013166
Annotation
PREDICTED:_histone_deacetylase_3_[Bombyx_mori]
Full name
Histone deacetylase
Location in the cell
Cytoplasmic Reliability : 1.218 Nuclear Reliability : 1.798
Sequence
CDS
ATGTGTCAACACTCGCCCGCAGTACAAATGACTCATATACCAGGTGACTTCTTCCCTGAAGATTACCGAATCAAGGACGAACCCGATCCTGACGTTCGTATCAGTCAGGACGATGCTGACAAAATGGTGGAGCCCAAAAACGAGTTTTACGAAGACGAGAAAGATAACGATAAGGAGTTGCCACCAGAGATTAAGGATCCATAG
Protein
MCQHSPAVQMTHIPGDFFPEDYRIKDEPDPDVRISQDDADKMVEPKNEFYEDEKDNDKELPPEIKDP
Summary
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Similarity
Belongs to the histone deacetylase family. HD Type 1 subfamily.
Belongs to the 14-3-3 family.
Belongs to the 14-3-3 family.
Uniprot
H9JUF3
A0A1E1WCE1
S4PV06
A0A2A4JPL6
A0A0B5WYL7
A0A2H1X2Q6
+ More
A0A437B1G7 A0A194PIE2 A0A212F234 A0A182VW75 A0A1L8DWY8 A0A1B0GIB0 A0A0N1I504 Q16QZ8 B0X1P4 A0A084W0K0 A0A023ERM0 A0A2M4BNS3 A0A1Y1K2Q7 A0A182HBL8 A0A2M3Z9Z7 A0A2M4ANK6 A0A182FSA0 W5J5F1 J3JTY3 A0A0A1WJN1 A0A2P8Y1E2 A0A0K8UZB8 A0A1L7N252 A0A0K8U3E5 A0A034WQK0 A0A336LLV5 W8CA23 A0A067R7R1 A0A182NQZ5 A0A1Q3F463 D6WYS7 A0A182QSK7 B4KD95 A0A182MEP8 B4LZC0 A0A182RG01 A0A182YNC5 A0A182SHD5 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A182XNZ7 A0A182PR77 A0A1A9XU26 A0A1A9VPK8 A0A1B0AVG5 A0A1A9ZJP7 A0A182JRG3 A0A2J7R7Y7 A0A1W4XTM3 B4JHV5 A0A3B0JQ91 B5DXY4 B4GF88 U5EVN6 A0A1B0GCY1 A0A0M5J4Z4 E0VKD4 A0A182IW81 A0A0L0BUJ3 B3LZH6 A0A0K8SU87 B4NB88 A0A1W4UJ00 A0A0A9WDV4 A0A2R7VRL2 Q7KTS4 Q9VNC2 B4PVA5 A0A1I8N7M5 B4QWW2 A0A2C9JHC5 B4I409 B3P2E0 A0A2R7VZD4 A0A2S2PJD5 T1GKI2 A0A1I8Q9N9 A0A3S1B0G7 A0A2T7NP68 A0A3R7T0P5 A0A3Q8Q043 A0A0C9R9J6 A0A2H8TMX3 J9KAR8 A0A0L8GDC4 A0A0B6YR45 A0A0D2WXJ4 A0A1B6GV27 A0A1B6JL65 A0A069DUM6 A0A210QF79
A0A437B1G7 A0A194PIE2 A0A212F234 A0A182VW75 A0A1L8DWY8 A0A1B0GIB0 A0A0N1I504 Q16QZ8 B0X1P4 A0A084W0K0 A0A023ERM0 A0A2M4BNS3 A0A1Y1K2Q7 A0A182HBL8 A0A2M3Z9Z7 A0A2M4ANK6 A0A182FSA0 W5J5F1 J3JTY3 A0A0A1WJN1 A0A2P8Y1E2 A0A0K8UZB8 A0A1L7N252 A0A0K8U3E5 A0A034WQK0 A0A336LLV5 W8CA23 A0A067R7R1 A0A182NQZ5 A0A1Q3F463 D6WYS7 A0A182QSK7 B4KD95 A0A182MEP8 B4LZC0 A0A182RG01 A0A182YNC5 A0A182SHD5 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A182XNZ7 A0A182PR77 A0A1A9XU26 A0A1A9VPK8 A0A1B0AVG5 A0A1A9ZJP7 A0A182JRG3 A0A2J7R7Y7 A0A1W4XTM3 B4JHV5 A0A3B0JQ91 B5DXY4 B4GF88 U5EVN6 A0A1B0GCY1 A0A0M5J4Z4 E0VKD4 A0A182IW81 A0A0L0BUJ3 B3LZH6 A0A0K8SU87 B4NB88 A0A1W4UJ00 A0A0A9WDV4 A0A2R7VRL2 Q7KTS4 Q9VNC2 B4PVA5 A0A1I8N7M5 B4QWW2 A0A2C9JHC5 B4I409 B3P2E0 A0A2R7VZD4 A0A2S2PJD5 T1GKI2 A0A1I8Q9N9 A0A3S1B0G7 A0A2T7NP68 A0A3R7T0P5 A0A3Q8Q043 A0A0C9R9J6 A0A2H8TMX3 J9KAR8 A0A0L8GDC4 A0A0B6YR45 A0A0D2WXJ4 A0A1B6GV27 A0A1B6JL65 A0A069DUM6 A0A210QF79
EC Number
3.5.1.98
Pubmed
19121390
23622113
28756777
26354079
22118469
17510324
+ More
24438588 24945155 28004739 26483478 20920257 23761445 22516182 23537049 25830018 29403074 27956627 25348373 24495485 24845553 18362917 19820115 17994087 18057021 25244985 20966253 12364791 14747013 17210077 15632085 20566863 26108605 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9852957 17550304 25315136 15562597 26334808 28812685
24438588 24945155 28004739 26483478 20920257 23761445 22516182 23537049 25830018 29403074 27956627 25348373 24495485 24845553 18362917 19820115 17994087 18057021 25244985 20966253 12364791 14747013 17210077 15632085 20566863 26108605 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9852957 17550304 25315136 15562597 26334808 28812685
EMBL
BABH01022812
GDQN01006404
JAT84650.1
GAIX01009373
JAA83187.1
NWSH01000876
+ More
PCG73759.1 KM983338 KZ150072 AJH76982.1 PZC74020.1 ODYU01013000 SOQ59595.1 RSAL01000212 RVE44174.1 KQ459602 KPI93181.1 AGBW02010783 OWR47805.1 GFDF01003121 JAV10963.1 AJWK01013622 AJWK01013623 KQ461178 KPJ07782.1 CH477727 EAT36822.1 DS232266 EDS38772.1 ATLV01019122 KE525262 KFB43744.1 GAPW01002272 JAC11326.1 GGFJ01005521 MBW54662.1 GEZM01098405 GEZM01098404 JAV53895.1 JXUM01125249 KQ566924 KXJ69730.1 GGFM01004590 MBW25341.1 GGFK01009055 MBW42376.1 ADMH02002066 ETN59692.1 APGK01019092 BT126693 KB740092 KB632429 AEE61655.1 ENN81439.1 ERL95457.1 GBXI01015572 JAC98719.1 PYGN01001050 PSN38082.1 GDHF01020310 GDHF01018819 JAI32004.1 JAI33495.1 LC096258 BAW19562.1 GDHF01031268 JAI21046.1 GAKP01002919 JAC56033.1 UFQT01000046 SSX18880.1 GAMC01001659 JAC04897.1 KK852685 KDR18505.1 KDR18506.1 GFDL01012701 JAV22344.1 KQ971357 EFA08457.1 AXCN02001485 CH933806 EDW14877.1 KRG01258.1 AXCM01000085 CH940650 EDW68155.1 AAAB01008987 EAA01056.5 APCN01000816 JXJN01004246 NEVH01006723 PNF36945.1 CH916369 EDV92863.1 OUUW01000008 SPP84367.1 CM000070 EDY68305.1 CH479182 EDW34273.1 GANO01001798 JAB58073.1 CCAG010002244 CP012526 ALC46648.1 DS235242 EEB13840.1 JRES01001304 KNC23745.1 CH902617 EDV41918.1 KPU79435.1 GBRD01008944 JAG56877.1 CH964232 EDW81052.1 GBHO01037002 GDHC01018870 JAG06602.1 JAP99758.1 KK854012 PTY09310.1 AE014297 AY061321 JQ663528 AAF52023.1 AAL28869.1 AFI26263.1 AF071559 AAC83649.1 CM000160 EDW95774.1 CM000364 EDX11724.1 CH480821 EDW54952.1 CH954181 EDV47890.1 KK854100 PTY11275.1 GGMR01016941 MBY29560.1 CAQQ02007907 CAQQ02007908 CAQQ02007909 RQTK01001150 RUS71853.1 PZQS01000010 PVD22970.1 QCYY01000543 ROT84428.1 MG491310 AZI71013.1 GBYB01004825 JAG74592.1 GFXV01003670 MBW15475.1 ABLF02027678 KQ422362 KOF75027.1 HACG01011757 CEK58622.1 KE346374 KJE97488.1 GECZ01003469 JAS66300.1 GECU01025655 GECU01007776 JAS82051.1 JAS99930.1 GBGD01001517 JAC87372.1 NEDP02003918 OWF47395.1
PCG73759.1 KM983338 KZ150072 AJH76982.1 PZC74020.1 ODYU01013000 SOQ59595.1 RSAL01000212 RVE44174.1 KQ459602 KPI93181.1 AGBW02010783 OWR47805.1 GFDF01003121 JAV10963.1 AJWK01013622 AJWK01013623 KQ461178 KPJ07782.1 CH477727 EAT36822.1 DS232266 EDS38772.1 ATLV01019122 KE525262 KFB43744.1 GAPW01002272 JAC11326.1 GGFJ01005521 MBW54662.1 GEZM01098405 GEZM01098404 JAV53895.1 JXUM01125249 KQ566924 KXJ69730.1 GGFM01004590 MBW25341.1 GGFK01009055 MBW42376.1 ADMH02002066 ETN59692.1 APGK01019092 BT126693 KB740092 KB632429 AEE61655.1 ENN81439.1 ERL95457.1 GBXI01015572 JAC98719.1 PYGN01001050 PSN38082.1 GDHF01020310 GDHF01018819 JAI32004.1 JAI33495.1 LC096258 BAW19562.1 GDHF01031268 JAI21046.1 GAKP01002919 JAC56033.1 UFQT01000046 SSX18880.1 GAMC01001659 JAC04897.1 KK852685 KDR18505.1 KDR18506.1 GFDL01012701 JAV22344.1 KQ971357 EFA08457.1 AXCN02001485 CH933806 EDW14877.1 KRG01258.1 AXCM01000085 CH940650 EDW68155.1 AAAB01008987 EAA01056.5 APCN01000816 JXJN01004246 NEVH01006723 PNF36945.1 CH916369 EDV92863.1 OUUW01000008 SPP84367.1 CM000070 EDY68305.1 CH479182 EDW34273.1 GANO01001798 JAB58073.1 CCAG010002244 CP012526 ALC46648.1 DS235242 EEB13840.1 JRES01001304 KNC23745.1 CH902617 EDV41918.1 KPU79435.1 GBRD01008944 JAG56877.1 CH964232 EDW81052.1 GBHO01037002 GDHC01018870 JAG06602.1 JAP99758.1 KK854012 PTY09310.1 AE014297 AY061321 JQ663528 AAF52023.1 AAL28869.1 AFI26263.1 AF071559 AAC83649.1 CM000160 EDW95774.1 CM000364 EDX11724.1 CH480821 EDW54952.1 CH954181 EDV47890.1 KK854100 PTY11275.1 GGMR01016941 MBY29560.1 CAQQ02007907 CAQQ02007908 CAQQ02007909 RQTK01001150 RUS71853.1 PZQS01000010 PVD22970.1 QCYY01000543 ROT84428.1 MG491310 AZI71013.1 GBYB01004825 JAG74592.1 GFXV01003670 MBW15475.1 ABLF02027678 KQ422362 KOF75027.1 HACG01011757 CEK58622.1 KE346374 KJE97488.1 GECZ01003469 JAS66300.1 GECU01025655 GECU01007776 JAS82051.1 JAS99930.1 GBGD01001517 JAC87372.1 NEDP02003918 OWF47395.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000075920
+ More
UP000092461 UP000053240 UP000008820 UP000002320 UP000030765 UP000069940 UP000249989 UP000069272 UP000000673 UP000019118 UP000030742 UP000245037 UP000027135 UP000075884 UP000007266 UP000075886 UP000009192 UP000075883 UP000008792 UP000075900 UP000076408 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075885 UP000092443 UP000078200 UP000092460 UP000092445 UP000075881 UP000235965 UP000192223 UP000001070 UP000268350 UP000001819 UP000008744 UP000092444 UP000092553 UP000009046 UP000075880 UP000037069 UP000007801 UP000007798 UP000192221 UP000000803 UP000002282 UP000095301 UP000000304 UP000076420 UP000001292 UP000008711 UP000015102 UP000095300 UP000271974 UP000245119 UP000283509 UP000007819 UP000053454 UP000008743 UP000242188
UP000092461 UP000053240 UP000008820 UP000002320 UP000030765 UP000069940 UP000249989 UP000069272 UP000000673 UP000019118 UP000030742 UP000245037 UP000027135 UP000075884 UP000007266 UP000075886 UP000009192 UP000075883 UP000008792 UP000075900 UP000076408 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075885 UP000092443 UP000078200 UP000092460 UP000092445 UP000075881 UP000235965 UP000192223 UP000001070 UP000268350 UP000001819 UP000008744 UP000092444 UP000092553 UP000009046 UP000075880 UP000037069 UP000007801 UP000007798 UP000192221 UP000000803 UP000002282 UP000095301 UP000000304 UP000076420 UP000001292 UP000008711 UP000015102 UP000095300 UP000271974 UP000245119 UP000283509 UP000007819 UP000053454 UP000008743 UP000242188
Interpro
Gene 3D
ProteinModelPortal
H9JUF3
A0A1E1WCE1
S4PV06
A0A2A4JPL6
A0A0B5WYL7
A0A2H1X2Q6
+ More
A0A437B1G7 A0A194PIE2 A0A212F234 A0A182VW75 A0A1L8DWY8 A0A1B0GIB0 A0A0N1I504 Q16QZ8 B0X1P4 A0A084W0K0 A0A023ERM0 A0A2M4BNS3 A0A1Y1K2Q7 A0A182HBL8 A0A2M3Z9Z7 A0A2M4ANK6 A0A182FSA0 W5J5F1 J3JTY3 A0A0A1WJN1 A0A2P8Y1E2 A0A0K8UZB8 A0A1L7N252 A0A0K8U3E5 A0A034WQK0 A0A336LLV5 W8CA23 A0A067R7R1 A0A182NQZ5 A0A1Q3F463 D6WYS7 A0A182QSK7 B4KD95 A0A182MEP8 B4LZC0 A0A182RG01 A0A182YNC5 A0A182SHD5 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A182XNZ7 A0A182PR77 A0A1A9XU26 A0A1A9VPK8 A0A1B0AVG5 A0A1A9ZJP7 A0A182JRG3 A0A2J7R7Y7 A0A1W4XTM3 B4JHV5 A0A3B0JQ91 B5DXY4 B4GF88 U5EVN6 A0A1B0GCY1 A0A0M5J4Z4 E0VKD4 A0A182IW81 A0A0L0BUJ3 B3LZH6 A0A0K8SU87 B4NB88 A0A1W4UJ00 A0A0A9WDV4 A0A2R7VRL2 Q7KTS4 Q9VNC2 B4PVA5 A0A1I8N7M5 B4QWW2 A0A2C9JHC5 B4I409 B3P2E0 A0A2R7VZD4 A0A2S2PJD5 T1GKI2 A0A1I8Q9N9 A0A3S1B0G7 A0A2T7NP68 A0A3R7T0P5 A0A3Q8Q043 A0A0C9R9J6 A0A2H8TMX3 J9KAR8 A0A0L8GDC4 A0A0B6YR45 A0A0D2WXJ4 A0A1B6GV27 A0A1B6JL65 A0A069DUM6 A0A210QF79
A0A437B1G7 A0A194PIE2 A0A212F234 A0A182VW75 A0A1L8DWY8 A0A1B0GIB0 A0A0N1I504 Q16QZ8 B0X1P4 A0A084W0K0 A0A023ERM0 A0A2M4BNS3 A0A1Y1K2Q7 A0A182HBL8 A0A2M3Z9Z7 A0A2M4ANK6 A0A182FSA0 W5J5F1 J3JTY3 A0A0A1WJN1 A0A2P8Y1E2 A0A0K8UZB8 A0A1L7N252 A0A0K8U3E5 A0A034WQK0 A0A336LLV5 W8CA23 A0A067R7R1 A0A182NQZ5 A0A1Q3F463 D6WYS7 A0A182QSK7 B4KD95 A0A182MEP8 B4LZC0 A0A182RG01 A0A182YNC5 A0A182SHD5 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A182XNZ7 A0A182PR77 A0A1A9XU26 A0A1A9VPK8 A0A1B0AVG5 A0A1A9ZJP7 A0A182JRG3 A0A2J7R7Y7 A0A1W4XTM3 B4JHV5 A0A3B0JQ91 B5DXY4 B4GF88 U5EVN6 A0A1B0GCY1 A0A0M5J4Z4 E0VKD4 A0A182IW81 A0A0L0BUJ3 B3LZH6 A0A0K8SU87 B4NB88 A0A1W4UJ00 A0A0A9WDV4 A0A2R7VRL2 Q7KTS4 Q9VNC2 B4PVA5 A0A1I8N7M5 B4QWW2 A0A2C9JHC5 B4I409 B3P2E0 A0A2R7VZD4 A0A2S2PJD5 T1GKI2 A0A1I8Q9N9 A0A3S1B0G7 A0A2T7NP68 A0A3R7T0P5 A0A3Q8Q043 A0A0C9R9J6 A0A2H8TMX3 J9KAR8 A0A0L8GDC4 A0A0B6YR45 A0A0D2WXJ4 A0A1B6GV27 A0A1B6JL65 A0A069DUM6 A0A210QF79
Ontologies
GO
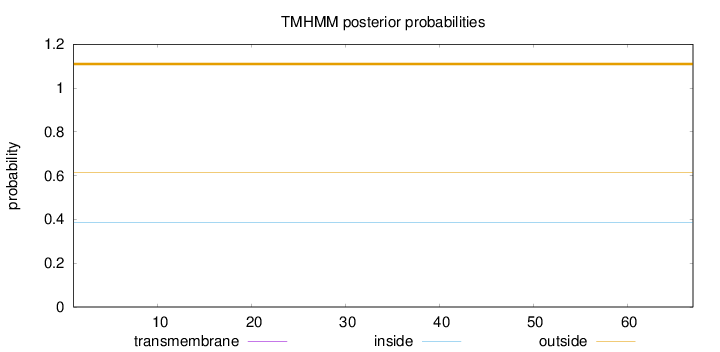
Topology
Subcellular location
Nucleus
Length:
67
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.38651
outside
1 - 67
Population Genetic Test Statistics
Pi
305.097701
Theta
157.504805
Tajima's D
3.141897
CLR
0.282439
CSRT
0.984300784960752
Interpretation
Uncertain
