Gene
KWMTBOMO09901
Pre Gene Modal
BGIBMGA013166
Annotation
PREDICTED:_histone_deacetylase_3_[Bombyx_mori]
Full name
Histone deacetylase
Location in the cell
Cytoplasmic Reliability : 1.12 Extracellular Reliability : 1.26
Sequence
CDS
ATGACTCAACAAAGGGTAGCGTATTTCTATAATCCGGACGTGGGTAATTTCCACTACGGTCCAGGGCATCCAATGAAACCACACAGGCTGGCTGTGACGCATTGTCTCGTGTTGAAGTATGGTCTTCATAAGAAAATGCAGATATATAAGCCGTATAGGGCCAGCGCTCACGATATGTGCCGTTTCCACAGCGAGGACTACATAGAGTTTCTGCAAAATGTAACTCCTCAAAATATACAGAGTTACTCAAAAGATCTTTTGCACTATAATGTTGGAGACGACTGTCCAGTATTCGAAGGTCTTTTCGATTTCTGCTCAATGTACACTGGAGCTTCATTGGAAGGTGCTATGAAGCTAAATAATAACACTTGTGACATTGCCATCAACTGGTCTGGGGGCTTGCATCATGCAAAGAAATTTGAACCTTCCGGATTTTGCTACGTCAATGATATAGTAATAGCAATTTTAGAGCTCCTAAAATATCATCCAAGGGTTCTGTACATAGATATTGATGTTCACCATGGAGATGGGGTTCAAGAAGCTTTTTACCTCACTGACCGAGTCATGACGGTTAGTTTCCACAAGTATGGGAACTATTTCTTCCCTGGAACGGGAGATATGTATGAAATTGGTGCAGAGAGTGGACGATATTATTCGGTAAATGTACCACTGAAAGAAGGCATAGATGATCAAAGCTACATACAGGTCAGTTTGGATGATTTATTATCTTAA
Protein
MTQQRVAYFYNPDVGNFHYGPGHPMKPHRLAVTHCLVLKYGLHKKMQIYKPYRASAHDMCRFHSEDYIEFLQNVTPQNIQSYSKDLLHYNVGDDCPVFEGLFDFCSMYTGASLEGAMKLNNNTCDIAINWSGGLHHAKKFEPSGFCYVNDIVIAILELLKYHPRVLYIDIDVHHGDGVQEAFYLTDRVMTVSFHKYGNYFFPGTGDMYEIGAESGRYYSVNVPLKEGIDDQSYIQVSLDDLLS
Summary
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Similarity
Belongs to the histone deacetylase family. HD Type 1 subfamily.
Belongs to the 14-3-3 family.
Belongs to the 14-3-3 family.
Uniprot
A0A437B1G7
S4PZV6
A0A212F234
A0A2H1X2Q6
A0A2A4JPL6
A0A0B5WYL7
+ More
A0A194PIE2 A0A2J7R7Y7 A0A067R7R1 A0A2P8Y1E2 A0A2A3ESH4 A0A0M8ZVL7 A0A087ZYB8 A0A0L7R0K4 E0VKD4 A0A154PHT9 E2BDQ7 A0A0L8FHS1 F4WLJ6 A0A195EDT5 A0A195AV22 A0A195FWD1 A0A158NN93 A0A151XAF7 A0A195CR97 E9IVN9 V5GPE9 A0A232EXV9 A0A0J7KJK1 E1ZX52 K7ILW2 A0A1W4XTM3 A0A1B0D7S5 A0A131XHG3 A0A026WVG4 A0A0V0G9K2 D6WYS7 A0A131Z5C7 A0A1Y1K2Q7 L7MDR1 A0A224YUX6 A0A023G4L6 A0A0P4VL73 V5HVX6 A0A069DUM6 A0A023FUV4 A0A023F5P1 A0A2L2Y721 A0A182XNZ7 A0A1Z5LFZ1 A0A182HBL8 Q16QZ8 T1HNM1 A0A1L7N252 U5EVN6 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A087UPN1 A0A182YNC5 A0A182IW81 A0A293M5N3 A0A182RG01 A0A0A9WDV4 T1J752 A0A182PR77 A0A182JRG3 A0A182MEP8 A0A182QSK7 A0A182NQZ5 A0A1B0GIB0 A0A2M4BNS3 A0A2M3Z9Z7 A0A182FSA0 W5J5F1 J9KAR8 A0A1L8DWY8 A0A084W0K0 A0A2H8TMX3 A0A2M4ANK6 A0A336LLV5 B0X1P4 A0A1E1XAS3 A0A2R7VRL2 A0A1Q3F463 A0A1D2MJ42 A0A0P5B0K4 A0A0P6FEZ1 A0A1S3IYF3 A0A0P6EVN2 E9GEV8 A0A0P6GFG8 A0A1B6LRK8 A0A0P5V7Q1 A0A0P5WXA5 A0A0P5EVU3 A0A210QF79 A0A1A9XU26
A0A194PIE2 A0A2J7R7Y7 A0A067R7R1 A0A2P8Y1E2 A0A2A3ESH4 A0A0M8ZVL7 A0A087ZYB8 A0A0L7R0K4 E0VKD4 A0A154PHT9 E2BDQ7 A0A0L8FHS1 F4WLJ6 A0A195EDT5 A0A195AV22 A0A195FWD1 A0A158NN93 A0A151XAF7 A0A195CR97 E9IVN9 V5GPE9 A0A232EXV9 A0A0J7KJK1 E1ZX52 K7ILW2 A0A1W4XTM3 A0A1B0D7S5 A0A131XHG3 A0A026WVG4 A0A0V0G9K2 D6WYS7 A0A131Z5C7 A0A1Y1K2Q7 L7MDR1 A0A224YUX6 A0A023G4L6 A0A0P4VL73 V5HVX6 A0A069DUM6 A0A023FUV4 A0A023F5P1 A0A2L2Y721 A0A182XNZ7 A0A1Z5LFZ1 A0A182HBL8 Q16QZ8 T1HNM1 A0A1L7N252 U5EVN6 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A087UPN1 A0A182YNC5 A0A182IW81 A0A293M5N3 A0A182RG01 A0A0A9WDV4 T1J752 A0A182PR77 A0A182JRG3 A0A182MEP8 A0A182QSK7 A0A182NQZ5 A0A1B0GIB0 A0A2M4BNS3 A0A2M3Z9Z7 A0A182FSA0 W5J5F1 J9KAR8 A0A1L8DWY8 A0A084W0K0 A0A2H8TMX3 A0A2M4ANK6 A0A336LLV5 B0X1P4 A0A1E1XAS3 A0A2R7VRL2 A0A1Q3F463 A0A1D2MJ42 A0A0P5B0K4 A0A0P6FEZ1 A0A1S3IYF3 A0A0P6EVN2 E9GEV8 A0A0P6GFG8 A0A1B6LRK8 A0A0P5V7Q1 A0A0P5WXA5 A0A0P5EVU3 A0A210QF79 A0A1A9XU26
EC Number
3.5.1.98
Pubmed
23622113
22118469
28756777
26354079
24845553
29403074
+ More
20566863 20798317 21719571 21347285 21282665 28648823 20075255 28049606 24508170 30249741 18362917 19820115 26830274 28004739 25576852 28797301 27129103 25765539 26334808 25474469 26561354 28528879 26483478 17510324 27956627 20966253 12364791 14747013 17210077 25244985 25401762 26823975 20920257 23761445 24438588 28503490 27289101 21292972 28812685
20566863 20798317 21719571 21347285 21282665 28648823 20075255 28049606 24508170 30249741 18362917 19820115 26830274 28004739 25576852 28797301 27129103 25765539 26334808 25474469 26561354 28528879 26483478 17510324 27956627 20966253 12364791 14747013 17210077 25244985 25401762 26823975 20920257 23761445 24438588 28503490 27289101 21292972 28812685
EMBL
RSAL01000212
RVE44174.1
GAIX01000873
JAA91687.1
AGBW02010783
OWR47805.1
+ More
ODYU01013000 SOQ59595.1 NWSH01000876 PCG73759.1 KM983338 KZ150072 AJH76982.1 PZC74020.1 KQ459602 KPI93181.1 NEVH01006723 PNF36945.1 KK852685 KDR18505.1 KDR18506.1 PYGN01001050 PSN38082.1 KZ288191 PBC34464.1 KQ435826 KOX72040.1 KQ414670 KOC64385.1 DS235242 EEB13840.1 KQ434912 KZC11393.1 GL447678 EFN86219.1 KQ431332 KOF63223.1 GL888208 EGI64932.1 KQ979039 KYN23368.1 KQ976738 KYM75875.1 KQ981208 KYN44753.1 ADTU01021179 KQ982351 KYQ57268.1 KQ977372 KYN03263.1 GL766328 EFZ15349.1 GALX01002482 JAB65984.1 NNAY01001656 OXU23306.1 LBMM01006632 KMQ90447.1 GL435014 EFN74242.1 AJVK01012514 GEFH01003505 JAP65076.1 KK107085 QOIP01000008 EZA60030.1 RLU19868.1 GECL01001493 JAP04631.1 KQ971357 EFA08457.1 GEDV01002911 JAP85646.1 GEZM01098405 GEZM01098404 JAV53895.1 GACK01002668 JAA62366.1 GFPF01008403 MAA19549.1 GBBM01006242 JAC29176.1 GDKW01000730 JAI55865.1 GANP01001704 JAB82764.1 GBGD01001517 JAC87372.1 GBBK01000029 JAC24453.1 GBBI01002228 JAC16484.1 IAAA01016117 LAA03969.1 GFJQ02000657 JAW06313.1 JXUM01125249 KQ566924 KXJ69730.1 CH477727 EAT36822.1 ACPB03012017 LC096258 BAW19562.1 GANO01001798 JAB58073.1 AAAB01008987 EAA01056.5 APCN01000816 KK120903 KFM79320.1 GFWV01009681 MAA34410.1 GBHO01037002 GDHC01018870 JAG06602.1 JAP99758.1 JH431912 AXCM01000085 AXCN02001485 AJWK01013622 AJWK01013623 GGFJ01005521 MBW54662.1 GGFM01004590 MBW25341.1 ADMH02002066 ETN59692.1 ABLF02027678 GFDF01003121 JAV10963.1 ATLV01019122 KE525262 KFB43744.1 GFXV01003670 MBW15475.1 GGFK01009055 MBW42376.1 UFQT01000046 SSX18880.1 DS232266 EDS38772.1 GFAC01002834 JAT96354.1 KK854012 PTY09310.1 GFDL01012701 JAV22344.1 LJIJ01001095 ODM93026.1 GDIP01191732 JAJ31670.1 GDIQ01060813 JAN33924.1 GDIQ01070157 JAN24580.1 GL732541 EFX81904.1 GDIQ01046326 JAN48411.1 GEBQ01013617 JAT26360.1 GDIP01104102 JAL99612.1 GDIP01080645 JAM23070.1 GDIP01233394 GDIP01233393 GDIP01154748 GDIP01154747 GDIP01148654 GDIP01148653 GDIQ01102898 GDIP01141854 GDIP01141853 GDIP01141852 GDIP01134453 GDIP01134452 GDIP01115204 GDIP01115203 GDIP01113758 GDIP01113757 GDIP01052264 GDIP01052263 GDIQ01062710 LRGB01002121 JAJ68654.1 JAL48828.1 KZS09068.1 NEDP02003918 OWF47395.1
ODYU01013000 SOQ59595.1 NWSH01000876 PCG73759.1 KM983338 KZ150072 AJH76982.1 PZC74020.1 KQ459602 KPI93181.1 NEVH01006723 PNF36945.1 KK852685 KDR18505.1 KDR18506.1 PYGN01001050 PSN38082.1 KZ288191 PBC34464.1 KQ435826 KOX72040.1 KQ414670 KOC64385.1 DS235242 EEB13840.1 KQ434912 KZC11393.1 GL447678 EFN86219.1 KQ431332 KOF63223.1 GL888208 EGI64932.1 KQ979039 KYN23368.1 KQ976738 KYM75875.1 KQ981208 KYN44753.1 ADTU01021179 KQ982351 KYQ57268.1 KQ977372 KYN03263.1 GL766328 EFZ15349.1 GALX01002482 JAB65984.1 NNAY01001656 OXU23306.1 LBMM01006632 KMQ90447.1 GL435014 EFN74242.1 AJVK01012514 GEFH01003505 JAP65076.1 KK107085 QOIP01000008 EZA60030.1 RLU19868.1 GECL01001493 JAP04631.1 KQ971357 EFA08457.1 GEDV01002911 JAP85646.1 GEZM01098405 GEZM01098404 JAV53895.1 GACK01002668 JAA62366.1 GFPF01008403 MAA19549.1 GBBM01006242 JAC29176.1 GDKW01000730 JAI55865.1 GANP01001704 JAB82764.1 GBGD01001517 JAC87372.1 GBBK01000029 JAC24453.1 GBBI01002228 JAC16484.1 IAAA01016117 LAA03969.1 GFJQ02000657 JAW06313.1 JXUM01125249 KQ566924 KXJ69730.1 CH477727 EAT36822.1 ACPB03012017 LC096258 BAW19562.1 GANO01001798 JAB58073.1 AAAB01008987 EAA01056.5 APCN01000816 KK120903 KFM79320.1 GFWV01009681 MAA34410.1 GBHO01037002 GDHC01018870 JAG06602.1 JAP99758.1 JH431912 AXCM01000085 AXCN02001485 AJWK01013622 AJWK01013623 GGFJ01005521 MBW54662.1 GGFM01004590 MBW25341.1 ADMH02002066 ETN59692.1 ABLF02027678 GFDF01003121 JAV10963.1 ATLV01019122 KE525262 KFB43744.1 GFXV01003670 MBW15475.1 GGFK01009055 MBW42376.1 UFQT01000046 SSX18880.1 DS232266 EDS38772.1 GFAC01002834 JAT96354.1 KK854012 PTY09310.1 GFDL01012701 JAV22344.1 LJIJ01001095 ODM93026.1 GDIP01191732 JAJ31670.1 GDIQ01060813 JAN33924.1 GDIQ01070157 JAN24580.1 GL732541 EFX81904.1 GDIQ01046326 JAN48411.1 GEBQ01013617 JAT26360.1 GDIP01104102 JAL99612.1 GDIP01080645 JAM23070.1 GDIP01233394 GDIP01233393 GDIP01154748 GDIP01154747 GDIP01148654 GDIP01148653 GDIQ01102898 GDIP01141854 GDIP01141853 GDIP01141852 GDIP01134453 GDIP01134452 GDIP01115204 GDIP01115203 GDIP01113758 GDIP01113757 GDIP01052264 GDIP01052263 GDIQ01062710 LRGB01002121 JAJ68654.1 JAL48828.1 KZS09068.1 NEDP02003918 OWF47395.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000235965
UP000027135
+ More
UP000245037 UP000242457 UP000053105 UP000005203 UP000053825 UP000009046 UP000076502 UP000008237 UP000053454 UP000007755 UP000078492 UP000078540 UP000078541 UP000005205 UP000075809 UP000078542 UP000215335 UP000036403 UP000000311 UP000002358 UP000192223 UP000092462 UP000053097 UP000279307 UP000007266 UP000076407 UP000069940 UP000249989 UP000008820 UP000015103 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000054359 UP000076408 UP000075880 UP000075900 UP000075885 UP000075881 UP000075883 UP000075886 UP000075884 UP000092461 UP000069272 UP000000673 UP000007819 UP000030765 UP000002320 UP000094527 UP000085678 UP000000305 UP000076858 UP000242188 UP000092443
UP000245037 UP000242457 UP000053105 UP000005203 UP000053825 UP000009046 UP000076502 UP000008237 UP000053454 UP000007755 UP000078492 UP000078540 UP000078541 UP000005205 UP000075809 UP000078542 UP000215335 UP000036403 UP000000311 UP000002358 UP000192223 UP000092462 UP000053097 UP000279307 UP000007266 UP000076407 UP000069940 UP000249989 UP000008820 UP000015103 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000054359 UP000076408 UP000075880 UP000075900 UP000075885 UP000075881 UP000075883 UP000075886 UP000075884 UP000092461 UP000069272 UP000000673 UP000007819 UP000030765 UP000002320 UP000094527 UP000085678 UP000000305 UP000076858 UP000242188 UP000092443
Interpro
Gene 3D
ProteinModelPortal
A0A437B1G7
S4PZV6
A0A212F234
A0A2H1X2Q6
A0A2A4JPL6
A0A0B5WYL7
+ More
A0A194PIE2 A0A2J7R7Y7 A0A067R7R1 A0A2P8Y1E2 A0A2A3ESH4 A0A0M8ZVL7 A0A087ZYB8 A0A0L7R0K4 E0VKD4 A0A154PHT9 E2BDQ7 A0A0L8FHS1 F4WLJ6 A0A195EDT5 A0A195AV22 A0A195FWD1 A0A158NN93 A0A151XAF7 A0A195CR97 E9IVN9 V5GPE9 A0A232EXV9 A0A0J7KJK1 E1ZX52 K7ILW2 A0A1W4XTM3 A0A1B0D7S5 A0A131XHG3 A0A026WVG4 A0A0V0G9K2 D6WYS7 A0A131Z5C7 A0A1Y1K2Q7 L7MDR1 A0A224YUX6 A0A023G4L6 A0A0P4VL73 V5HVX6 A0A069DUM6 A0A023FUV4 A0A023F5P1 A0A2L2Y721 A0A182XNZ7 A0A1Z5LFZ1 A0A182HBL8 Q16QZ8 T1HNM1 A0A1L7N252 U5EVN6 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A087UPN1 A0A182YNC5 A0A182IW81 A0A293M5N3 A0A182RG01 A0A0A9WDV4 T1J752 A0A182PR77 A0A182JRG3 A0A182MEP8 A0A182QSK7 A0A182NQZ5 A0A1B0GIB0 A0A2M4BNS3 A0A2M3Z9Z7 A0A182FSA0 W5J5F1 J9KAR8 A0A1L8DWY8 A0A084W0K0 A0A2H8TMX3 A0A2M4ANK6 A0A336LLV5 B0X1P4 A0A1E1XAS3 A0A2R7VRL2 A0A1Q3F463 A0A1D2MJ42 A0A0P5B0K4 A0A0P6FEZ1 A0A1S3IYF3 A0A0P6EVN2 E9GEV8 A0A0P6GFG8 A0A1B6LRK8 A0A0P5V7Q1 A0A0P5WXA5 A0A0P5EVU3 A0A210QF79 A0A1A9XU26
A0A194PIE2 A0A2J7R7Y7 A0A067R7R1 A0A2P8Y1E2 A0A2A3ESH4 A0A0M8ZVL7 A0A087ZYB8 A0A0L7R0K4 E0VKD4 A0A154PHT9 E2BDQ7 A0A0L8FHS1 F4WLJ6 A0A195EDT5 A0A195AV22 A0A195FWD1 A0A158NN93 A0A151XAF7 A0A195CR97 E9IVN9 V5GPE9 A0A232EXV9 A0A0J7KJK1 E1ZX52 K7ILW2 A0A1W4XTM3 A0A1B0D7S5 A0A131XHG3 A0A026WVG4 A0A0V0G9K2 D6WYS7 A0A131Z5C7 A0A1Y1K2Q7 L7MDR1 A0A224YUX6 A0A023G4L6 A0A0P4VL73 V5HVX6 A0A069DUM6 A0A023FUV4 A0A023F5P1 A0A2L2Y721 A0A182XNZ7 A0A1Z5LFZ1 A0A182HBL8 Q16QZ8 T1HNM1 A0A1L7N252 U5EVN6 A0A182VGB9 A0A182TF68 A0A182L8D0 Q7PX00 A0A182HK42 A0A087UPN1 A0A182YNC5 A0A182IW81 A0A293M5N3 A0A182RG01 A0A0A9WDV4 T1J752 A0A182PR77 A0A182JRG3 A0A182MEP8 A0A182QSK7 A0A182NQZ5 A0A1B0GIB0 A0A2M4BNS3 A0A2M3Z9Z7 A0A182FSA0 W5J5F1 J9KAR8 A0A1L8DWY8 A0A084W0K0 A0A2H8TMX3 A0A2M4ANK6 A0A336LLV5 B0X1P4 A0A1E1XAS3 A0A2R7VRL2 A0A1Q3F463 A0A1D2MJ42 A0A0P5B0K4 A0A0P6FEZ1 A0A1S3IYF3 A0A0P6EVN2 E9GEV8 A0A0P6GFG8 A0A1B6LRK8 A0A0P5V7Q1 A0A0P5WXA5 A0A0P5EVU3 A0A210QF79 A0A1A9XU26
PDB
4A69
E-value=2.68791e-114,
Score=1051
Ontologies
GO
Topology
Subcellular location
Nucleus
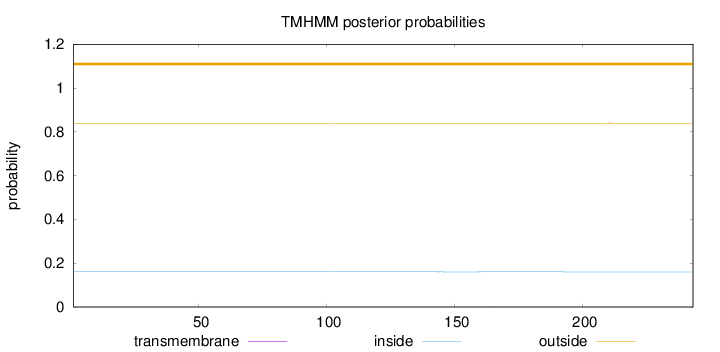
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02426
Exp number, first 60 AAs:
0.00492
Total prob of N-in:
0.16085
outside
1 - 243
Population Genetic Test Statistics
Pi
14.499429
Theta
18.230547
Tajima's D
-0.72985
CLR
0.749271
CSRT
0.191190440477976
Interpretation
Uncertain
