Gene
KWMTBOMO09898
Pre Gene Modal
BGIBMGA013165
Annotation
PREDICTED:_cell_cycle_checkpoint_protein_RAD1_[Amyelois_transitella]
Full name
Cell cycle checkpoint protein RAD1
Alternative Name
DNA repair exonuclease rad1 homolog
Rad1-like DNA damage checkpoint protein
Rad1-like DNA damage checkpoint protein
Location in the cell
Cytoplasmic Reliability : 1.75 Extracellular Reliability : 1.799
Sequence
CDS
ATGGCTGAAGATTCATCTCAATACCATTTCACGGCTAGCATGGACTCCGGAAAGACTCTGTGTAGTTTATTAAAAGCAATACAGTTTCAAGAGTCAGCAGTGTTCTGTGCTCTACCTGAAGGTTTGAAGTTAACAGTGGAGGAAGGCAAGTGTGTTCAAGCCTCAGCTTATATACCGGCTGACAACTTCACAGAATACCATGTTAGGGATGATGTAGATGTCATATTTAAGATCAGCTTAGCAGTTTTAGCAGAATGCTTAAACATATTTGGCTCGGGTGAAGAGGCCAGCTTGAAAATGTATTACAAATGTGAAGGATCCCCGCTGTTACTTGTCTTACAGCCTCAGTCGTTTCGTAACGTCATGACGGACTGCGAGATATGGAGCCAGACCCCAGATTCGATGCTGGAGCTTCGCAGTCCCGAGGAGACGGACGTGGCGAAGCTCGTGGTCCGCGCCGGAGCCCTACTCGCAGTACTGGGGGACGTCGAGAGGAGTGCCGATGTCTTGGAGTTGAGGTTGCAGCCTGAACCGCCACACTTCACCATCGTTACGTACGGAATGCAGGATCGATCCTGTATCGAAGTGCCTGAGTGCTCGGACACACTGCAGAGCTTCAGCTGTGAACGAGAACTGACCCTCAAATATCAACTCCACCACCTCAGGCTCTGCATGAAAGCTCTAGCTATTTCAAATAAGGTCGTATTAAGATGCAGCTCCACGGGCCTGTTATTGCTTCAGCTGAAGTTAGAAAGGGACGACAACAAACACATGTTCTCTGAATTCTATATAGTGCCTCTACTTGACGACTGA
Protein
MAEDSSQYHFTASMDSGKTLCSLLKAIQFQESAVFCALPEGLKLTVEEGKCVQASAYIPADNFTEYHVRDDVDVIFKISLAVLAECLNIFGSGEEASLKMYYKCEGSPLLLVLQPQSFRNVMTDCEIWSQTPDSMLELRSPEETDVAKLVVRAGALLAVLGDVERSADVLELRLQPEPPHFTIVTYGMQDRSCIEVPECSDTLQSFSCERELTLKYQLHHLRLCMKALAISNKVVLRCSSTGLLLLQLKLERDDNKHMFSEFYIVPLLDD
Summary
Description
Component of the 9-1-1 cell-cycle checkpoint response complex that plays a major role in DNA repair. The 9-1-1 complex is recruited to DNA lesion upon damage by the RAD17-replication factor C (RFC) clamp loader complex. Acts then as a sliding clamp platform on DNA for several proteins involved in long-patch base excision repair (LP-BER). The 9-1-1 complex stimulates DNA polymerase beta (POLB) activity by increasing its affinity for the 3'-OH end of the primer-template and stabilizes POLB to those sites where LP-BER proceeds; endonuclease FEN1 cleavage activity on substrates with double, nick, or gap flaps of distinct sequences and lengths; and DNA ligase I (LIG1) on long-patch base excision repair substrates. The 9-1-1 complex is necessary for the recruitment of RHNO1 to sites of double-stranded breaks (DSB) occurring during the S phase. Isoform 1 possesses 3'->5' double stranded DNA exonuclease activity (By similarity).
Catalytic Activity
Exonucleolytic cleavage in the 3'- to 5'-direction to yield nucleoside 5'-phosphates.
Subunit
Component of the toroidal 9-1-1 (RAD9-RAD1-HUS1) complex, composed of RAD9A, RAD1 and HUS1. The 9-1-1 complex associates with LIG1, POLB, FEN1, RAD17, HDAC1, RPA1 and RPA2. The 9-1-1 complex associates with the RAD17-RFC complex. RAD1 interacts with POLB, FEN1, HUS1, HUS1B, RAD9A and RAD9B. Interacts with DNAJC7 (By similarity).
Similarity
Belongs to the rad1 family.
Keywords
Alternative splicing
Complete proteome
DNA damage
DNA repair
Exonuclease
Hydrolase
Nuclease
Nucleus
Reference proteome
Feature
chain Cell cycle checkpoint protein RAD1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A1E1W2F6
A0A2H1VY32
A0A2A4J6D1
S4NMZ8
A0A437ARY1
A0A212F278
+ More
A0A0N1IDA7 A0A194PIQ4 D6X3Z8 A0A1W4WCV2 A0A3Q4AAP1 A0A154P6B5 A0A3Q1IQ40 A0A3P8WWP6 A0A0L8FK96 A0A226PI72 C1JXM2 A7T5Z1 A0A210Q0J5 A0A088AEC1 A0A3B4WYG8 A0A1D5PSK5 A0A1S3K1Z2 A0A1B6DL23 A0A3B5KJE7 A0A3B4BJ66 A0A3Q1GS54 G3PVY0 A0A3Q1B7P3 A0A3P8TH84 A0A2A3ERR4 A0A2U9CFL8 A0A2U9CKW4 A0A3B4UWK8 A0A232FJV2 C3XV97 A0A3Q3FXG3 A0A3P9HTH5 H2LMW7 A0A3P9K7N5 A0A3B4ZIR0 A0A2J7Q1I2 A0A0F8CAP2 W8B5C0 A0A3M0LBQ3 U3JHD7 A0A2Z5U607 A0A034W1M4 A0A444U5D4 A7SU20 H0YUP7 A0A3L8S8T6 A0A1V4KDN1 I3JLK1 A0A3Q3MMV4 R7V578 A0A3Q3CVX6 A0A3Q3K8P5 K7IZC6 A0A2I0LR64 A0A0B6ZTE3 Q9QWZ1 A0A0B6ZRK9 A0A1A7WEU2 A0A067QQH9 A0A3P8NSE4 A0A3P9CW37 A0A3B3YYM5 A0A087XSC6 A0A3B3TWE6 A0A2I4BAI1 H2YAI1 A0A3Q2EA67 G3HKF5 A9ULD8 A0A3P9PVL8 A0A099Z1L3 A0A3L7I509 A0A250YEE3 A0A1U8C7T6 D3ZC52 A0A2I0UBD2 A0A091LYM9 C1BZ21 A0A3B4FK40 A0A218UVJ5 Q7ZWD7 A0A3Q3ARC2 Q6P2T4 A0A147A229 A0A3B4BVV1 A0A1A8RBS4 A0A060YHT7 A0A3Q2Q9K7 A0A1A8QW88 A0A1S3FCS8 A0A093QFT4
A0A0N1IDA7 A0A194PIQ4 D6X3Z8 A0A1W4WCV2 A0A3Q4AAP1 A0A154P6B5 A0A3Q1IQ40 A0A3P8WWP6 A0A0L8FK96 A0A226PI72 C1JXM2 A7T5Z1 A0A210Q0J5 A0A088AEC1 A0A3B4WYG8 A0A1D5PSK5 A0A1S3K1Z2 A0A1B6DL23 A0A3B5KJE7 A0A3B4BJ66 A0A3Q1GS54 G3PVY0 A0A3Q1B7P3 A0A3P8TH84 A0A2A3ERR4 A0A2U9CFL8 A0A2U9CKW4 A0A3B4UWK8 A0A232FJV2 C3XV97 A0A3Q3FXG3 A0A3P9HTH5 H2LMW7 A0A3P9K7N5 A0A3B4ZIR0 A0A2J7Q1I2 A0A0F8CAP2 W8B5C0 A0A3M0LBQ3 U3JHD7 A0A2Z5U607 A0A034W1M4 A0A444U5D4 A7SU20 H0YUP7 A0A3L8S8T6 A0A1V4KDN1 I3JLK1 A0A3Q3MMV4 R7V578 A0A3Q3CVX6 A0A3Q3K8P5 K7IZC6 A0A2I0LR64 A0A0B6ZTE3 Q9QWZ1 A0A0B6ZRK9 A0A1A7WEU2 A0A067QQH9 A0A3P8NSE4 A0A3P9CW37 A0A3B3YYM5 A0A087XSC6 A0A3B3TWE6 A0A2I4BAI1 H2YAI1 A0A3Q2EA67 G3HKF5 A9ULD8 A0A3P9PVL8 A0A099Z1L3 A0A3L7I509 A0A250YEE3 A0A1U8C7T6 D3ZC52 A0A2I0UBD2 A0A091LYM9 C1BZ21 A0A3B4FK40 A0A218UVJ5 Q7ZWD7 A0A3Q3ARC2 Q6P2T4 A0A147A229 A0A3B4BVV1 A0A1A8RBS4 A0A060YHT7 A0A3Q2Q9K7 A0A1A8QW88 A0A1S3FCS8 A0A093QFT4
EC Number
3.1.11.2
Pubmed
23622113
22118469
26354079
18362917
19820115
24487278
+ More
24621616 17615350 28812685 15592404 21551351 25463417 28648823 18563158 17554307 25835551 24495485 26760975 25348373 20360741 30282656 25186727 23254933 20075255 23371554 9716408 9828137 9878245 9660799 9705507 16141072 15489334 24845553 21804562 20431018 29704459 28087693 15057822 15632090 20433749 25069045 23594743 24755649
24621616 17615350 28812685 15592404 21551351 25463417 28648823 18563158 17554307 25835551 24495485 26760975 25348373 20360741 30282656 25186727 23254933 20075255 23371554 9716408 9828137 9878245 9660799 9705507 16141072 15489334 24845553 21804562 20431018 29704459 28087693 15057822 15632090 20433749 25069045 23594743 24755649
EMBL
GDQN01011450
GDQN01009898
GDQN01005752
GDQN01004168
GDQN01001002
JAT79604.1
+ More
JAT81156.1 JAT85302.1 JAT86886.1 JAT90052.1 ODYU01005159 SOQ45750.1 NWSH01002689 PCG67677.1 GAIX01014161 JAA78399.1 RSAL01001726 RVE40723.1 AGBW02010783 OWR47837.1 KQ461178 KPJ07761.1 KQ459602 KPI93202.1 KQ971379 EEZ97345.1 KQ434827 KZC07469.1 KQ429968 KOF64876.1 AWGT02000073 OXB79159.1 FJ705223 ACO52484.1 DS471358 EDO28618.1 NEDP02005302 OWF42256.1 AC192388 GEDC01010914 JAS26384.1 KZ288192 PBC34397.1 CP026258 AWP15391.1 AWP15392.1 NNAY01000112 OXU30863.1 GG666468 EEN68023.1 NEVH01019409 PNF22435.1 KQ042382 KKF15334.1 GAMC01010215 JAB96340.1 QRBI01000102 RMC16607.1 AGTO01011146 FX985771 BBA93658.1 GAKP01009481 GAKP01009480 JAC49471.1 SCEB01215283 RXM30402.1 DS469806 EDO32797.1 ABQF01020732 QUSF01000044 RLV98173.1 LSYS01003582 OPJ82463.1 AERX01026660 AMQN01000734 KB295062 ELU13679.1 AKCR02000130 PKK19919.1 HACG01024120 CEK70985.1 AF074718 AF073523 AF076842 AJ004976 AF038841 AF058394 AF084514 AK146533 AK148098 AK150466 AK168588 BC048693 HACG01024122 CEK70987.1 HADW01002845 HADX01011989 SBP04245.1 KK853042 KDR12148.1 AYCK01007596 JH000458 EGV92308.1 AAMC01097948 BC157217 AAI57218.1 KL888260 KGL75611.1 RAZU01000096 RLQ73298.1 GFFW01002790 JAV41998.1 AC128789 CH474058 EDL82989.1 KZ505910 PKU43354.1 KK515829 KFP64708.1 BT079850 ACO14274.1 MUZQ01000116 OWK57793.1 BC049464 AAH49464.1 CR848814 BC064305 AAH64305.1 GCES01013712 JAR72611.1 HAEI01006551 HAEH01015810 SBS03525.1 FR908876 CDQ88690.1 HAEG01014319 SBR97284.1 KL693574 KFW87803.1
JAT81156.1 JAT85302.1 JAT86886.1 JAT90052.1 ODYU01005159 SOQ45750.1 NWSH01002689 PCG67677.1 GAIX01014161 JAA78399.1 RSAL01001726 RVE40723.1 AGBW02010783 OWR47837.1 KQ461178 KPJ07761.1 KQ459602 KPI93202.1 KQ971379 EEZ97345.1 KQ434827 KZC07469.1 KQ429968 KOF64876.1 AWGT02000073 OXB79159.1 FJ705223 ACO52484.1 DS471358 EDO28618.1 NEDP02005302 OWF42256.1 AC192388 GEDC01010914 JAS26384.1 KZ288192 PBC34397.1 CP026258 AWP15391.1 AWP15392.1 NNAY01000112 OXU30863.1 GG666468 EEN68023.1 NEVH01019409 PNF22435.1 KQ042382 KKF15334.1 GAMC01010215 JAB96340.1 QRBI01000102 RMC16607.1 AGTO01011146 FX985771 BBA93658.1 GAKP01009481 GAKP01009480 JAC49471.1 SCEB01215283 RXM30402.1 DS469806 EDO32797.1 ABQF01020732 QUSF01000044 RLV98173.1 LSYS01003582 OPJ82463.1 AERX01026660 AMQN01000734 KB295062 ELU13679.1 AKCR02000130 PKK19919.1 HACG01024120 CEK70985.1 AF074718 AF073523 AF076842 AJ004976 AF038841 AF058394 AF084514 AK146533 AK148098 AK150466 AK168588 BC048693 HACG01024122 CEK70987.1 HADW01002845 HADX01011989 SBP04245.1 KK853042 KDR12148.1 AYCK01007596 JH000458 EGV92308.1 AAMC01097948 BC157217 AAI57218.1 KL888260 KGL75611.1 RAZU01000096 RLQ73298.1 GFFW01002790 JAV41998.1 AC128789 CH474058 EDL82989.1 KZ505910 PKU43354.1 KK515829 KFP64708.1 BT079850 ACO14274.1 MUZQ01000116 OWK57793.1 BC049464 AAH49464.1 CR848814 BC064305 AAH64305.1 GCES01013712 JAR72611.1 HAEI01006551 HAEH01015810 SBS03525.1 FR908876 CDQ88690.1 HAEG01014319 SBR97284.1 KL693574 KFW87803.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000192223 UP000261620 UP000076502 UP000265040 UP000265120 UP000053454 UP000198419 UP000001593 UP000242188 UP000005203 UP000261360 UP000000539 UP000085678 UP000005226 UP000261520 UP000257200 UP000007635 UP000257160 UP000265080 UP000242457 UP000246464 UP000261420 UP000215335 UP000001554 UP000261660 UP000265200 UP000001038 UP000265180 UP000261400 UP000235965 UP000269221 UP000016665 UP000007754 UP000276834 UP000190648 UP000005207 UP000261640 UP000014760 UP000264840 UP000261600 UP000002358 UP000053872 UP000000589 UP000027135 UP000265100 UP000265160 UP000261480 UP000028760 UP000261500 UP000192220 UP000007875 UP000265020 UP000001075 UP000008143 UP000242638 UP000053641 UP000273346 UP000189706 UP000002494 UP000265140 UP000261460 UP000197619 UP000264800 UP000000437 UP000261440 UP000193380 UP000265000 UP000081671 UP000053258
UP000192223 UP000261620 UP000076502 UP000265040 UP000265120 UP000053454 UP000198419 UP000001593 UP000242188 UP000005203 UP000261360 UP000000539 UP000085678 UP000005226 UP000261520 UP000257200 UP000007635 UP000257160 UP000265080 UP000242457 UP000246464 UP000261420 UP000215335 UP000001554 UP000261660 UP000265200 UP000001038 UP000265180 UP000261400 UP000235965 UP000269221 UP000016665 UP000007754 UP000276834 UP000190648 UP000005207 UP000261640 UP000014760 UP000264840 UP000261600 UP000002358 UP000053872 UP000000589 UP000027135 UP000265100 UP000265160 UP000261480 UP000028760 UP000261500 UP000192220 UP000007875 UP000265020 UP000001075 UP000008143 UP000242638 UP000053641 UP000273346 UP000189706 UP000002494 UP000265140 UP000261460 UP000197619 UP000264800 UP000000437 UP000261440 UP000193380 UP000265000 UP000081671 UP000053258
Pfam
PF02144 Rad1
ProteinModelPortal
A0A1E1W2F6
A0A2H1VY32
A0A2A4J6D1
S4NMZ8
A0A437ARY1
A0A212F278
+ More
A0A0N1IDA7 A0A194PIQ4 D6X3Z8 A0A1W4WCV2 A0A3Q4AAP1 A0A154P6B5 A0A3Q1IQ40 A0A3P8WWP6 A0A0L8FK96 A0A226PI72 C1JXM2 A7T5Z1 A0A210Q0J5 A0A088AEC1 A0A3B4WYG8 A0A1D5PSK5 A0A1S3K1Z2 A0A1B6DL23 A0A3B5KJE7 A0A3B4BJ66 A0A3Q1GS54 G3PVY0 A0A3Q1B7P3 A0A3P8TH84 A0A2A3ERR4 A0A2U9CFL8 A0A2U9CKW4 A0A3B4UWK8 A0A232FJV2 C3XV97 A0A3Q3FXG3 A0A3P9HTH5 H2LMW7 A0A3P9K7N5 A0A3B4ZIR0 A0A2J7Q1I2 A0A0F8CAP2 W8B5C0 A0A3M0LBQ3 U3JHD7 A0A2Z5U607 A0A034W1M4 A0A444U5D4 A7SU20 H0YUP7 A0A3L8S8T6 A0A1V4KDN1 I3JLK1 A0A3Q3MMV4 R7V578 A0A3Q3CVX6 A0A3Q3K8P5 K7IZC6 A0A2I0LR64 A0A0B6ZTE3 Q9QWZ1 A0A0B6ZRK9 A0A1A7WEU2 A0A067QQH9 A0A3P8NSE4 A0A3P9CW37 A0A3B3YYM5 A0A087XSC6 A0A3B3TWE6 A0A2I4BAI1 H2YAI1 A0A3Q2EA67 G3HKF5 A9ULD8 A0A3P9PVL8 A0A099Z1L3 A0A3L7I509 A0A250YEE3 A0A1U8C7T6 D3ZC52 A0A2I0UBD2 A0A091LYM9 C1BZ21 A0A3B4FK40 A0A218UVJ5 Q7ZWD7 A0A3Q3ARC2 Q6P2T4 A0A147A229 A0A3B4BVV1 A0A1A8RBS4 A0A060YHT7 A0A3Q2Q9K7 A0A1A8QW88 A0A1S3FCS8 A0A093QFT4
A0A0N1IDA7 A0A194PIQ4 D6X3Z8 A0A1W4WCV2 A0A3Q4AAP1 A0A154P6B5 A0A3Q1IQ40 A0A3P8WWP6 A0A0L8FK96 A0A226PI72 C1JXM2 A7T5Z1 A0A210Q0J5 A0A088AEC1 A0A3B4WYG8 A0A1D5PSK5 A0A1S3K1Z2 A0A1B6DL23 A0A3B5KJE7 A0A3B4BJ66 A0A3Q1GS54 G3PVY0 A0A3Q1B7P3 A0A3P8TH84 A0A2A3ERR4 A0A2U9CFL8 A0A2U9CKW4 A0A3B4UWK8 A0A232FJV2 C3XV97 A0A3Q3FXG3 A0A3P9HTH5 H2LMW7 A0A3P9K7N5 A0A3B4ZIR0 A0A2J7Q1I2 A0A0F8CAP2 W8B5C0 A0A3M0LBQ3 U3JHD7 A0A2Z5U607 A0A034W1M4 A0A444U5D4 A7SU20 H0YUP7 A0A3L8S8T6 A0A1V4KDN1 I3JLK1 A0A3Q3MMV4 R7V578 A0A3Q3CVX6 A0A3Q3K8P5 K7IZC6 A0A2I0LR64 A0A0B6ZTE3 Q9QWZ1 A0A0B6ZRK9 A0A1A7WEU2 A0A067QQH9 A0A3P8NSE4 A0A3P9CW37 A0A3B3YYM5 A0A087XSC6 A0A3B3TWE6 A0A2I4BAI1 H2YAI1 A0A3Q2EA67 G3HKF5 A9ULD8 A0A3P9PVL8 A0A099Z1L3 A0A3L7I509 A0A250YEE3 A0A1U8C7T6 D3ZC52 A0A2I0UBD2 A0A091LYM9 C1BZ21 A0A3B4FK40 A0A218UVJ5 Q7ZWD7 A0A3Q3ARC2 Q6P2T4 A0A147A229 A0A3B4BVV1 A0A1A8RBS4 A0A060YHT7 A0A3Q2Q9K7 A0A1A8QW88 A0A1S3FCS8 A0A093QFT4
PDB
3A1J
E-value=5.37625e-34,
Score=359
Ontologies
GO
PANTHER
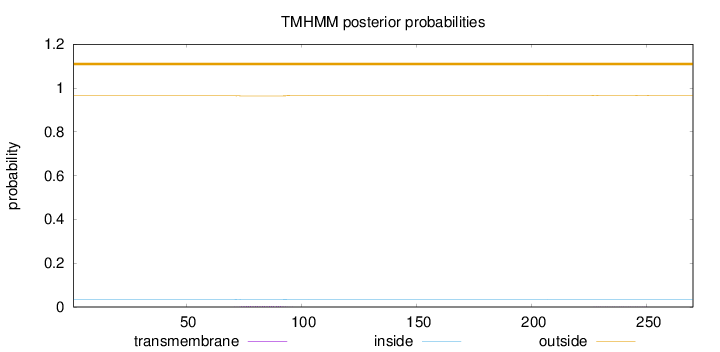
Topology
Subcellular location
Nucleus
Length:
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09307
Exp number, first 60 AAs:
0.00111
Total prob of N-in:
0.03504
outside
1 - 270
Population Genetic Test Statistics
Pi
248.214437
Theta
152.489647
Tajima's D
2.642859
CLR
0.114002
CSRT
0.954752262386881
Interpretation
Uncertain
