Gene
KWMTBOMO09897
Pre Gene Modal
BGIBMGA013263
Annotation
PREDICTED:_solute_carrier_family_35_member_G1_[Bombyx_mori]
Full name
1-acyl-sn-glycerol-3-phosphate acyltransferase
Location in the cell
PlasmaMembrane Reliability : 4.993
Sequence
CDS
ATGCCAGAACATGTGGAACTTCAACAACTCGTGGATATGTCAGTCGGCAGCGGGGATGAGTCCATCCCCAATCTACTCGAGAACAAGAGACCTCTTATAAAGCGATGTCCATATCTTGGTTTGATTTTAGCTACTTTATCCTCTCTATTTTTCTCACTATGTTCAGTTATAGTAAAGAGTTTAGTCAACATAGATCCAATGCAACTGGCAATGTTTCGGTTCATCGGAGTACTTTTACCGACGATACCTATAGTTGTATACACCGAGCAGCCAGTCTTTCCACAAGGTAAACGTTTATTGCTAGCTTTACGTTCCATTGTCGGAACAGTAGGTCTCATGCTGAGTTTTTACGCATTCAGGAATATGCCGCTAGCAGATGCATCCGTCATAGTGTTCTCTGTGCCCGTTTTCGTAGCACTTTTCGCGCGAGTTTTTTTAAAAGAACCCTGCGGTATGTGGAATACAATTTCCATAATTCTTACGTTAATTGGTGTTATATTAATAACACATCCGCCATTTTTATTTGGTGGTACAGTAACAGAACATAATTCAAACTATAACAGTCTTAGAGGAGCTGTAGCTGCATTTATATCGACAATATTTGGAGCAAATGCCTATGTTCTTCTAAGAGTACTCAAAGGATTACATTTTTCAGTGATCATGACAAATTTTGGAGCTATAGCAATTTTACAAACATTTGTCTATTCATTTGTTTTTGGCTTGCTATGCATGCCAAATTGTGGCACAGAACGTTTCCTTGTTGTTTGTTTGGCTTTGTTTAGTTATTTGGGTCAGATCTTATTAACAATGTCATTACAATTGGAACAGGCAGGCCCTGTGGCCATAGCTCGATCAGCTGATATTGTTTTCGCCTTTCTATGGCAGGTGATGTTTTTTAATGAAATCCCAAGTAAATTTTCAATTTGCGGTGCTGTTTTAGTGTTGAGTTCTGTTATGTTAGTTGGTTTAAGGAAATGGGCGTTAGCTCTACCGGAAGACTCTAGTTTAAGGAGTAAATTAGGTGCACTTGCACGATAA
Protein
MPEHVELQQLVDMSVGSGDESIPNLLENKRPLIKRCPYLGLILATLSSLFFSLCSVIVKSLVNIDPMQLAMFRFIGVLLPTIPIVVYTEQPVFPQGKRLLLALRSIVGTVGLMLSFYAFRNMPLADASVIVFSVPVFVALFARVFLKEPCGMWNTISIILTLIGVILITHPPFLFGGTVTEHNSNYNSLRGAVAAFISTIFGANAYVLLRVLKGLHFSVIMTNFGAIAILQTFVYSFVFGLLCMPNCGTERFLVVCLALFSYLGQILLTMSLQLEQAGPVAIARSADIVFAFLWQVMFFNEIPSKFSICGAVLVLSSVMLVGLRKWALALPEDSSLRSKLGALAR
Summary
Catalytic Activity
a 1-acyl-sn-glycero-3-phosphate + an acyl-CoA = a 1,2-diacyl-sn-glycero-3-phosphate + CoA
Similarity
Belongs to the 1-acyl-sn-glycerol-3-phosphate acyltransferase family.
Uniprot
H9JUQ0
A0A2A4J7N9
A0A0L7LB86
S4P8Y7
A0A212F233
A0A0N0PAR9
+ More
A0A194PIE7 K7J0W1 A0A232FF52 A0A158NVT1 A0A195EDA1 F4WNR5 A0A195B8I1 A0A195FVQ1 D2A414 E9J5U0 A0A151IP33 E0VMP0 W5J6U6 A0A182FS35 A0A182YHW7 A0A1L8DF66 A0A1L8DF15 A0A3L8E2R4 A0A026WG90 A0A0L7RG84 A0A182QQD1 A0A182V4M6 A0A182TT81 A0A182X9C2 A0A182L8I6 Q7PX50 A0A182HKB0 A0A182SVF5 A0A087ZVP8 A0A182RSE0 A0A084W021 A0A182JKH5 A0A182NQS7 A0A182M4L0 A0A182PRW0 A0A154PIC0 E2BRH3 A0A182VW12 A0A2A3EA69 B4KDZ1 A0A310S8T2 A0A2J7Q6U8 B4NAW9 A0A1B0GCU7 A0A1B0A491 A0A182GJG4 A0A1B0C4P1 A0A1A9XU72 A0A1I8NXX0 A0A1A9VR78 B4M5C9 E2B266 A0A151XBW8 B0WK59 A0A0A1WHW7 W8B0A4 A0A0K8V9H9 A0A1S4FPH3 Q16TQ5 A0A1W4W1E5 B3NZE6 A0A1I8MKJ1 B4HI23 B4QU35 Q9VGN6 A0A3B0KIQ1 B4JUP0 A0A1B6E1D0 A0A1B6HFV8 B4PM59 Q295A3 A0A0M4ESW3 N6SXU5 A0A067RDB2 B3M0X0 A0A023F223 A0A034WC64 A0A1W4WMF0 A0A2R7VS54 U4U4C8 T1HU02 A0A224XUJ7 B4GMD6 A0A0M9A5G2 J9JY59 A0A336LQ09 A0A336LKJ9 A0A2P8YB78 A0A1W4WCC4 A0A2S2N8R8 E9G8N5 A0A0N8D7Y8 T1J654 A0A0P5TKC8
A0A194PIE7 K7J0W1 A0A232FF52 A0A158NVT1 A0A195EDA1 F4WNR5 A0A195B8I1 A0A195FVQ1 D2A414 E9J5U0 A0A151IP33 E0VMP0 W5J6U6 A0A182FS35 A0A182YHW7 A0A1L8DF66 A0A1L8DF15 A0A3L8E2R4 A0A026WG90 A0A0L7RG84 A0A182QQD1 A0A182V4M6 A0A182TT81 A0A182X9C2 A0A182L8I6 Q7PX50 A0A182HKB0 A0A182SVF5 A0A087ZVP8 A0A182RSE0 A0A084W021 A0A182JKH5 A0A182NQS7 A0A182M4L0 A0A182PRW0 A0A154PIC0 E2BRH3 A0A182VW12 A0A2A3EA69 B4KDZ1 A0A310S8T2 A0A2J7Q6U8 B4NAW9 A0A1B0GCU7 A0A1B0A491 A0A182GJG4 A0A1B0C4P1 A0A1A9XU72 A0A1I8NXX0 A0A1A9VR78 B4M5C9 E2B266 A0A151XBW8 B0WK59 A0A0A1WHW7 W8B0A4 A0A0K8V9H9 A0A1S4FPH3 Q16TQ5 A0A1W4W1E5 B3NZE6 A0A1I8MKJ1 B4HI23 B4QU35 Q9VGN6 A0A3B0KIQ1 B4JUP0 A0A1B6E1D0 A0A1B6HFV8 B4PM59 Q295A3 A0A0M4ESW3 N6SXU5 A0A067RDB2 B3M0X0 A0A023F223 A0A034WC64 A0A1W4WMF0 A0A2R7VS54 U4U4C8 T1HU02 A0A224XUJ7 B4GMD6 A0A0M9A5G2 J9JY59 A0A336LQ09 A0A336LKJ9 A0A2P8YB78 A0A1W4WCC4 A0A2S2N8R8 E9G8N5 A0A0N8D7Y8 T1J654 A0A0P5TKC8
EC Number
2.3.1.51
Pubmed
19121390
26227816
23622113
22118469
26354079
20075255
+ More
28648823 21347285 21719571 18362917 19820115 21282665 20566863 20920257 23761445 25244985 30249741 24508170 20966253 12364791 14747013 17210077 24438588 20798317 17994087 26483478 18057021 25830018 24495485 17510324 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23537049 24845553 25474469 25348373 29403074 21292972
28648823 21347285 21719571 18362917 19820115 21282665 20566863 20920257 23761445 25244985 30249741 24508170 20966253 12364791 14747013 17210077 24438588 20798317 17994087 26483478 18057021 25830018 24495485 17510324 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23537049 24845553 25474469 25348373 29403074 21292972
EMBL
BABH01022814
NWSH01002689
PCG67678.1
JTDY01001882
KOB72644.1
GAIX01003974
+ More
JAA88586.1 AGBW02010783 OWR47800.1 KQ461178 KPJ07777.1 KQ459602 KPI93186.1 NNAY01000332 OXU29150.1 ADTU01002902 KQ979074 KYN22797.1 GL888237 EGI64258.1 KQ976565 KYM80550.1 KQ981215 KYN44501.1 KQ971348 EFA05598.1 GL768196 EFZ11813.1 KQ976892 KYN07237.1 DS235321 EEB14646.1 ADMH02002101 ETN59123.1 GFDF01009057 JAV05027.1 GFDF01009056 JAV05028.1 QOIP01000001 RLU27031.1 KK107260 EZA54059.1 KQ414599 KOC69756.1 AXCN02001500 AAAB01008987 EAA01799.1 APCN01000824 ATLV01019081 KE525262 KFB43565.1 AXCM01006044 KQ434924 KZC11605.1 GL449965 EFN81665.1 KZ288327 PBC27961.1 CH933806 EDW16012.1 KQ769895 OAD52771.1 NEVH01017450 PNF24309.1 CH964232 EDW80933.1 CCAG010002232 JXUM01068183 KQ562487 KXJ75801.1 JXJN01025564 CH940652 EDW59840.1 KRF79099.1 KRF79100.1 GL445059 EFN60241.1 KQ982316 KYQ57854.1 DS231968 EDS29633.1 GBXI01015835 JAC98456.1 GAMC01012046 GAMC01012045 JAB94510.1 GDHF01028865 GDHF01016871 JAI23449.1 JAI35443.1 CH477643 EAT37873.1 CH954181 EDV49519.1 CH480815 EDW42605.1 CM000364 EDX13366.1 AE014297 BT050556 AAF54640.1 ACJ13263.1 OUUW01000014 SPP88400.1 CH916374 EDV91210.1 GEDC01005566 JAS31732.1 GECU01034205 JAS73501.1 CM000160 EDW96925.1 CM000070 EAL28809.1 CP012526 ALC45805.1 APGK01052956 KB741216 ENN72564.1 KK852575 KDR21024.1 CH902617 EDV43199.1 KPU80127.1 GBBI01003095 JAC15617.1 GAKP01007559 JAC51393.1 KK854052 PTY10347.1 KB632003 ERL87897.1 ACPB03024218 GFTR01004777 JAW11649.1 CH479185 EDW38010.1 KQ435750 KOX76249.1 ABLF02018956 UFQT01000042 SSX18723.1 SSX18724.1 PYGN01000736 PSN41500.1 GGMR01000911 MBY13530.1 GL732535 EFX84236.1 GDIP01059943 GDIQ01042723 LRGB01002451 JAM43772.1 JAN52014.1 KZS07551.1 JH431872 GDIP01126605 JAL77109.1
JAA88586.1 AGBW02010783 OWR47800.1 KQ461178 KPJ07777.1 KQ459602 KPI93186.1 NNAY01000332 OXU29150.1 ADTU01002902 KQ979074 KYN22797.1 GL888237 EGI64258.1 KQ976565 KYM80550.1 KQ981215 KYN44501.1 KQ971348 EFA05598.1 GL768196 EFZ11813.1 KQ976892 KYN07237.1 DS235321 EEB14646.1 ADMH02002101 ETN59123.1 GFDF01009057 JAV05027.1 GFDF01009056 JAV05028.1 QOIP01000001 RLU27031.1 KK107260 EZA54059.1 KQ414599 KOC69756.1 AXCN02001500 AAAB01008987 EAA01799.1 APCN01000824 ATLV01019081 KE525262 KFB43565.1 AXCM01006044 KQ434924 KZC11605.1 GL449965 EFN81665.1 KZ288327 PBC27961.1 CH933806 EDW16012.1 KQ769895 OAD52771.1 NEVH01017450 PNF24309.1 CH964232 EDW80933.1 CCAG010002232 JXUM01068183 KQ562487 KXJ75801.1 JXJN01025564 CH940652 EDW59840.1 KRF79099.1 KRF79100.1 GL445059 EFN60241.1 KQ982316 KYQ57854.1 DS231968 EDS29633.1 GBXI01015835 JAC98456.1 GAMC01012046 GAMC01012045 JAB94510.1 GDHF01028865 GDHF01016871 JAI23449.1 JAI35443.1 CH477643 EAT37873.1 CH954181 EDV49519.1 CH480815 EDW42605.1 CM000364 EDX13366.1 AE014297 BT050556 AAF54640.1 ACJ13263.1 OUUW01000014 SPP88400.1 CH916374 EDV91210.1 GEDC01005566 JAS31732.1 GECU01034205 JAS73501.1 CM000160 EDW96925.1 CM000070 EAL28809.1 CP012526 ALC45805.1 APGK01052956 KB741216 ENN72564.1 KK852575 KDR21024.1 CH902617 EDV43199.1 KPU80127.1 GBBI01003095 JAC15617.1 GAKP01007559 JAC51393.1 KK854052 PTY10347.1 KB632003 ERL87897.1 ACPB03024218 GFTR01004777 JAW11649.1 CH479185 EDW38010.1 KQ435750 KOX76249.1 ABLF02018956 UFQT01000042 SSX18723.1 SSX18724.1 PYGN01000736 PSN41500.1 GGMR01000911 MBY13530.1 GL732535 EFX84236.1 GDIP01059943 GDIQ01042723 LRGB01002451 JAM43772.1 JAN52014.1 KZS07551.1 JH431872 GDIP01126605 JAL77109.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000002358 UP000215335 UP000005205 UP000078492 UP000007755 UP000078540 UP000078541 UP000007266 UP000078542 UP000009046 UP000000673 UP000069272 UP000076408 UP000279307 UP000053097 UP000053825 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075901 UP000005203 UP000075900 UP000030765 UP000075880 UP000075884 UP000075883 UP000075885 UP000076502 UP000008237 UP000075920 UP000242457 UP000009192 UP000235965 UP000007798 UP000092444 UP000092445 UP000069940 UP000249989 UP000092460 UP000092443 UP000095300 UP000078200 UP000008792 UP000000311 UP000075809 UP000002320 UP000008820 UP000192221 UP000008711 UP000095301 UP000001292 UP000000304 UP000000803 UP000268350 UP000001070 UP000002282 UP000001819 UP000092553 UP000019118 UP000027135 UP000007801 UP000192223 UP000030742 UP000015103 UP000008744 UP000053105 UP000007819 UP000245037 UP000000305 UP000076858
UP000002358 UP000215335 UP000005205 UP000078492 UP000007755 UP000078540 UP000078541 UP000007266 UP000078542 UP000009046 UP000000673 UP000069272 UP000076408 UP000279307 UP000053097 UP000053825 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075901 UP000005203 UP000075900 UP000030765 UP000075880 UP000075884 UP000075883 UP000075885 UP000076502 UP000008237 UP000075920 UP000242457 UP000009192 UP000235965 UP000007798 UP000092444 UP000092445 UP000069940 UP000249989 UP000092460 UP000092443 UP000095300 UP000078200 UP000008792 UP000000311 UP000075809 UP000002320 UP000008820 UP000192221 UP000008711 UP000095301 UP000001292 UP000000304 UP000000803 UP000268350 UP000001070 UP000002282 UP000001819 UP000092553 UP000019118 UP000027135 UP000007801 UP000192223 UP000030742 UP000015103 UP000008744 UP000053105 UP000007819 UP000245037 UP000000305 UP000076858
Pfam
Interpro
IPR000620
EamA_dom
+ More
IPR003124 WH2_dom
IPR000210 BTB/POZ_dom
IPR000465 XPA
IPR002123 Plipid/glycerol_acylTrfase
IPR019786 Zinc_finger_PHD-type_CS
IPR011333 SKP1/BTB/POZ_sf
IPR037129 XPA_sf
IPR020846 MFS_dom
IPR004552 AGP_acyltrans
IPR022656 XPA_C
IPR029147 CFAP77
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR009061 DNA-bd_dom_put_sf
IPR013083 Znf_RING/FYVE/PHD
IPR022652 Znf_XPA_CS
IPR019787 Znf_PHD-finger
IPR003131 T1-type_BTB
IPR003124 WH2_dom
IPR000210 BTB/POZ_dom
IPR000465 XPA
IPR002123 Plipid/glycerol_acylTrfase
IPR019786 Zinc_finger_PHD-type_CS
IPR011333 SKP1/BTB/POZ_sf
IPR037129 XPA_sf
IPR020846 MFS_dom
IPR004552 AGP_acyltrans
IPR022656 XPA_C
IPR029147 CFAP77
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR009061 DNA-bd_dom_put_sf
IPR013083 Znf_RING/FYVE/PHD
IPR022652 Znf_XPA_CS
IPR019787 Znf_PHD-finger
IPR003131 T1-type_BTB
Gene 3D
CDD
ProteinModelPortal
H9JUQ0
A0A2A4J7N9
A0A0L7LB86
S4P8Y7
A0A212F233
A0A0N0PAR9
+ More
A0A194PIE7 K7J0W1 A0A232FF52 A0A158NVT1 A0A195EDA1 F4WNR5 A0A195B8I1 A0A195FVQ1 D2A414 E9J5U0 A0A151IP33 E0VMP0 W5J6U6 A0A182FS35 A0A182YHW7 A0A1L8DF66 A0A1L8DF15 A0A3L8E2R4 A0A026WG90 A0A0L7RG84 A0A182QQD1 A0A182V4M6 A0A182TT81 A0A182X9C2 A0A182L8I6 Q7PX50 A0A182HKB0 A0A182SVF5 A0A087ZVP8 A0A182RSE0 A0A084W021 A0A182JKH5 A0A182NQS7 A0A182M4L0 A0A182PRW0 A0A154PIC0 E2BRH3 A0A182VW12 A0A2A3EA69 B4KDZ1 A0A310S8T2 A0A2J7Q6U8 B4NAW9 A0A1B0GCU7 A0A1B0A491 A0A182GJG4 A0A1B0C4P1 A0A1A9XU72 A0A1I8NXX0 A0A1A9VR78 B4M5C9 E2B266 A0A151XBW8 B0WK59 A0A0A1WHW7 W8B0A4 A0A0K8V9H9 A0A1S4FPH3 Q16TQ5 A0A1W4W1E5 B3NZE6 A0A1I8MKJ1 B4HI23 B4QU35 Q9VGN6 A0A3B0KIQ1 B4JUP0 A0A1B6E1D0 A0A1B6HFV8 B4PM59 Q295A3 A0A0M4ESW3 N6SXU5 A0A067RDB2 B3M0X0 A0A023F223 A0A034WC64 A0A1W4WMF0 A0A2R7VS54 U4U4C8 T1HU02 A0A224XUJ7 B4GMD6 A0A0M9A5G2 J9JY59 A0A336LQ09 A0A336LKJ9 A0A2P8YB78 A0A1W4WCC4 A0A2S2N8R8 E9G8N5 A0A0N8D7Y8 T1J654 A0A0P5TKC8
A0A194PIE7 K7J0W1 A0A232FF52 A0A158NVT1 A0A195EDA1 F4WNR5 A0A195B8I1 A0A195FVQ1 D2A414 E9J5U0 A0A151IP33 E0VMP0 W5J6U6 A0A182FS35 A0A182YHW7 A0A1L8DF66 A0A1L8DF15 A0A3L8E2R4 A0A026WG90 A0A0L7RG84 A0A182QQD1 A0A182V4M6 A0A182TT81 A0A182X9C2 A0A182L8I6 Q7PX50 A0A182HKB0 A0A182SVF5 A0A087ZVP8 A0A182RSE0 A0A084W021 A0A182JKH5 A0A182NQS7 A0A182M4L0 A0A182PRW0 A0A154PIC0 E2BRH3 A0A182VW12 A0A2A3EA69 B4KDZ1 A0A310S8T2 A0A2J7Q6U8 B4NAW9 A0A1B0GCU7 A0A1B0A491 A0A182GJG4 A0A1B0C4P1 A0A1A9XU72 A0A1I8NXX0 A0A1A9VR78 B4M5C9 E2B266 A0A151XBW8 B0WK59 A0A0A1WHW7 W8B0A4 A0A0K8V9H9 A0A1S4FPH3 Q16TQ5 A0A1W4W1E5 B3NZE6 A0A1I8MKJ1 B4HI23 B4QU35 Q9VGN6 A0A3B0KIQ1 B4JUP0 A0A1B6E1D0 A0A1B6HFV8 B4PM59 Q295A3 A0A0M4ESW3 N6SXU5 A0A067RDB2 B3M0X0 A0A023F223 A0A034WC64 A0A1W4WMF0 A0A2R7VS54 U4U4C8 T1HU02 A0A224XUJ7 B4GMD6 A0A0M9A5G2 J9JY59 A0A336LQ09 A0A336LKJ9 A0A2P8YB78 A0A1W4WCC4 A0A2S2N8R8 E9G8N5 A0A0N8D7Y8 T1J654 A0A0P5TKC8
Ontologies
GO
PANTHER
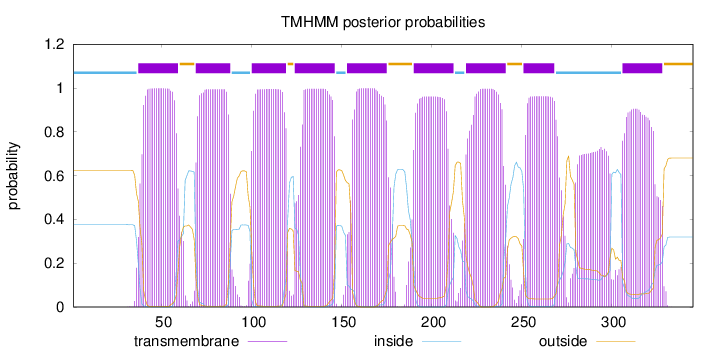
Topology
Length:
345
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
201.27146
Exp number, first 60 AAs:
21.30614
Total prob of N-in:
0.37648
POSSIBLE N-term signal
sequence
inside
1 - 36
TMhelix
37 - 59
outside
60 - 68
TMhelix
69 - 88
inside
89 - 99
TMhelix
100 - 119
outside
120 - 123
TMhelix
124 - 146
inside
147 - 152
TMhelix
153 - 175
outside
176 - 189
TMhelix
190 - 212
inside
213 - 218
TMhelix
219 - 241
outside
242 - 250
TMhelix
251 - 268
inside
269 - 305
TMhelix
306 - 328
outside
329 - 345
Population Genetic Test Statistics
Pi
294.782529
Theta
230.75663
Tajima's D
0.837909
CLR
1.329879
CSRT
0.615269236538173
Interpretation
Uncertain
