Gene
KWMTBOMO09892 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013267
Annotation
PREDICTED:_phosphatidylethanolamine-binding_protein_homolog_F40A3.3-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.211 Extracellular Reliability : 1.045
Sequence
CDS
ATGTCTTTGGTAGCACAAGCATTCGAAGCTAGCAAAATAGTGCCGGATGTTATTCCCGTCGCTCCAACTAAGAACATTGAGCTTAAATACCCGAGCGGTGCTATAGCGTCTCAAGGAAATGAGCTGACTCCGACACAAGTGAAGGATCAGCCTTCTGTCACTTTTGAGGCCGAGGCTGATGCTTTTTATACGCTCGTGTTCACGGACCCTGATAACTACGATGGACCTGAGCTGGTCTACCGTGAATGGCACCACTGGCTCGTAGGAAACATTCCTGGTGGTGACGTTTCTGCTGGAGAGACTCTATCGGGATACATCGGCTCCGGACCACCCCAGGGTACTGGGATCCATCGTTATGTCTACATCCTTTATAAACAACCAGGAAAATTGGACTTCGACGAGAAAAGGCTAACGAACACGTCTATTGATGGTCGCGCCTCGTTCTCCACGAAGAAGTTCGCTGAAAAGTACAACCTGGGGGCGCCTGTCGCCGGCAACTTCTACAGAGCGCAGTTCGACGACTATGTCCCACAGCTTTACAAAAGCTTGGGCGCCTAA
Protein
MSLVAQAFEASKIVPDVIPVAPTKNIELKYPSGAIASQGNELTPTQVKDQPSVTFEAEADAFYTLVFTDPDNYDGPELVYREWHHWLVGNIPGGDVSAGETLSGYIGSGPPQGTGIHRYVYILYKQPGKLDFDEKRLTNTSIDGRASFSTKKFAEKYNLGAPVAGNFYRAQFDDYVPQLYKSLGA
Summary
Uniprot
H9JUQ4
A0A2H1VKW9
A0A2H1X2U3
A0A3S2PKG7
A0A212F259
A0A194PJH0
+ More
I4DNG3 A0A2M3YWB1 A0A2M3Z7V4 A0A182F1E0 W5J4A4 A0A2M4A958 Q1HPY7 Q1HPY8 T1DN25 A0A2M3Z9F2 Q1HQT8 Q16LF6 A0A2M4BZN8 I4DM94 A0A084VL08 S4PMB7 I4DIX6 A0A194PIN5 A0A023EIJ5 A0A023ELU7 A0A0K8TS78 A0A023EJG8 A0A2H8TK93 A0A2S2QI33 B0WRQ3 C4WTE3 U5EQ08 T1P8H0 A0A1I8MNV8 C4WTE2 A0A2H1X2Y7 A0A182IZ08 U4U508 A0A212F257 N6TQS9 A0A1Q3FE74 A0A1L8DR19 A0A1Y1NCM5 A0A1W4X0J4 A0A2W1BIB2 A0A2S2NX04 A0A1L8DR75 J9JYE5 A0A1L8DRE0 A0A1I8Q1V0 A0A2A4JPX5 A0A1W4XB58 A0A1B0DH84 A0A1W4WJY1 C4WUE6 T1DIR4 A0A0A1WTC4 A0A1Y1NCL9 A0A182RSJ7 A0A1L8EBW4 A0A1L8EBV7 A0A1L8EB45 A0A1L8EC88 A0A1L8EBW5 A0A182P8B1 A0A182QV86 W8C068 A0A182I7A8 A0A219YQA9 A0A0L7KU41 A0A219YQ51 A0A411G7T8 A0A0K8UQL9 A0A232F887 A0A182UQ84 A0A182TEW0 Q7PWN1 A0A034WAM2 A0A182JUM3 A0A182XNP9 A0A182Y0B3 V5GYS1 A0A219YR16 A0A034WE87 A0A0C9QE70 A0A182LMV6 A0A182MJ69 K7IVL8 A0A182MZ75 A0A437B228 E9IP20 F4WAF3 K7IVM0 A0A2H8U169 A0A0N0BJX0 A0A158NKV7 A0A182VUD7 A0A336MQ06 E2ABH4
I4DNG3 A0A2M3YWB1 A0A2M3Z7V4 A0A182F1E0 W5J4A4 A0A2M4A958 Q1HPY7 Q1HPY8 T1DN25 A0A2M3Z9F2 Q1HQT8 Q16LF6 A0A2M4BZN8 I4DM94 A0A084VL08 S4PMB7 I4DIX6 A0A194PIN5 A0A023EIJ5 A0A023ELU7 A0A0K8TS78 A0A023EJG8 A0A2H8TK93 A0A2S2QI33 B0WRQ3 C4WTE3 U5EQ08 T1P8H0 A0A1I8MNV8 C4WTE2 A0A2H1X2Y7 A0A182IZ08 U4U508 A0A212F257 N6TQS9 A0A1Q3FE74 A0A1L8DR19 A0A1Y1NCM5 A0A1W4X0J4 A0A2W1BIB2 A0A2S2NX04 A0A1L8DR75 J9JYE5 A0A1L8DRE0 A0A1I8Q1V0 A0A2A4JPX5 A0A1W4XB58 A0A1B0DH84 A0A1W4WJY1 C4WUE6 T1DIR4 A0A0A1WTC4 A0A1Y1NCL9 A0A182RSJ7 A0A1L8EBW4 A0A1L8EBV7 A0A1L8EB45 A0A1L8EC88 A0A1L8EBW5 A0A182P8B1 A0A182QV86 W8C068 A0A182I7A8 A0A219YQA9 A0A0L7KU41 A0A219YQ51 A0A411G7T8 A0A0K8UQL9 A0A232F887 A0A182UQ84 A0A182TEW0 Q7PWN1 A0A034WAM2 A0A182JUM3 A0A182XNP9 A0A182Y0B3 V5GYS1 A0A219YR16 A0A034WE87 A0A0C9QE70 A0A182LMV6 A0A182MJ69 K7IVL8 A0A182MZ75 A0A437B228 E9IP20 F4WAF3 K7IVM0 A0A2H8U169 A0A0N0BJX0 A0A158NKV7 A0A182VUD7 A0A336MQ06 E2ABH4
Pubmed
19121390
22118469
26354079
22651552
20920257
23761445
+ More
17204158 17510324 24438588 23622113 24945155 26483478 26369729 25315136 23537049 28004739 28756777 24330624 25830018 24495485 28647560 26227816 28648823 12364791 14747013 17210077 25348373 25244985 20966253 20075255 21282665 21719571 21347285 20798317
17204158 17510324 24438588 23622113 24945155 26483478 26369729 25315136 23537049 28004739 28756777 24330624 25830018 24495485 28647560 26227816 28648823 12364791 14747013 17210077 25348373 25244985 20966253 20075255 21282665 21719571 21347285 20798317
EMBL
BABH01022822
BABH01022823
BABH01022824
ODYU01003120
SOQ41480.1
ODYU01013000
+ More
SOQ59597.1 RSAL01000008 RVE53995.1 AGBW02010783 OWR47803.1 KQ459602 KPI93183.1 AK402894 BAM19453.1 GGFF01000085 MBW20552.1 GGFM01003767 MBW24518.1 ADMH02002157 ETN58218.1 GGFK01004015 MBW37336.1 DQ443265 ABF51354.1 BABH01022813 DQ443264 ABF51353.1 GAMD01003235 JAA98355.1 GGFM01004393 MBW25144.1 DQ440356 CH477447 ABF18389.1 EAT40755.1 CH477908 EAT35142.1 GGFJ01009406 MBW58547.1 AK402412 BAM19034.1 ATLV01014395 KE524970 KFB38652.1 GAIX01003650 JAA88910.1 AK401244 BAM17866.1 KPI93182.1 JXUM01159252 GAPW01004355 KQ573291 JAC09243.1 KXJ67942.1 GAPW01004354 GAPW01004353 JAC09244.1 JAC09245.1 GDAI01000602 JAI17001.1 GAPW01004493 JAC09105.1 GFXV01002586 MBW14391.1 GGMS01007629 MBY76832.1 DS232058 EDS33457.1 ABLF02039519 AK340578 BAH71163.1 GANO01003484 JAB56387.1 KA645031 AFP59660.1 AK340577 BAH71162.1 SOQ59596.1 KB632130 ERL88969.1 OWR47804.1 APGK01024357 APGK01024358 KB740544 ENN80418.1 GFDL01009191 JAV25854.1 GFDF01005172 JAV08912.1 GEZM01006615 GEZM01006614 JAV95684.1 KZ150072 PZC74021.1 GGMR01009076 MBY21695.1 GFDF01005173 JAV08911.1 ABLF02039514 GFDF01005174 JAV08910.1 NWSH01000876 PCG73758.1 AJVK01060758 ABLF02039511 ABLF02039515 ABLF02039538 AK341032 BAH71516.1 GALA01000821 JAA94031.1 GBXI01012537 JAD01755.1 GEZM01006616 JAV95682.1 GFDG01002568 JAV16231.1 GFDG01002571 JAV16228.1 GFDG01002974 JAV15825.1 GFDG01002570 JAV16229.1 GFDG01002569 JAV16230.1 AXCN02002069 GAMC01004142 JAC02414.1 APCN01003375 KX270974 AQT33307.1 JTDY01005653 KOB66777.1 KX270975 AQT33309.1 MH366181 QBB01990.1 GDHF01023526 JAI28788.1 NNAY01000752 OXU26688.1 AAAB01008984 EAA14862.4 GAKP01006331 JAC52621.1 GALX01002873 JAB65593.1 KX270976 AQT33308.1 GAKP01006330 JAC52622.1 GBYB01012698 JAG82465.1 AXCM01000345 RSAL01000212 RVE44175.1 GL764397 EFZ17714.1 GL888048 EGI68836.1 GFXV01007777 MBW19582.1 KQ435710 KOX79734.1 ADTU01018970 UFQT01001398 SSX30377.1 GL438248 EFN69213.1
SOQ59597.1 RSAL01000008 RVE53995.1 AGBW02010783 OWR47803.1 KQ459602 KPI93183.1 AK402894 BAM19453.1 GGFF01000085 MBW20552.1 GGFM01003767 MBW24518.1 ADMH02002157 ETN58218.1 GGFK01004015 MBW37336.1 DQ443265 ABF51354.1 BABH01022813 DQ443264 ABF51353.1 GAMD01003235 JAA98355.1 GGFM01004393 MBW25144.1 DQ440356 CH477447 ABF18389.1 EAT40755.1 CH477908 EAT35142.1 GGFJ01009406 MBW58547.1 AK402412 BAM19034.1 ATLV01014395 KE524970 KFB38652.1 GAIX01003650 JAA88910.1 AK401244 BAM17866.1 KPI93182.1 JXUM01159252 GAPW01004355 KQ573291 JAC09243.1 KXJ67942.1 GAPW01004354 GAPW01004353 JAC09244.1 JAC09245.1 GDAI01000602 JAI17001.1 GAPW01004493 JAC09105.1 GFXV01002586 MBW14391.1 GGMS01007629 MBY76832.1 DS232058 EDS33457.1 ABLF02039519 AK340578 BAH71163.1 GANO01003484 JAB56387.1 KA645031 AFP59660.1 AK340577 BAH71162.1 SOQ59596.1 KB632130 ERL88969.1 OWR47804.1 APGK01024357 APGK01024358 KB740544 ENN80418.1 GFDL01009191 JAV25854.1 GFDF01005172 JAV08912.1 GEZM01006615 GEZM01006614 JAV95684.1 KZ150072 PZC74021.1 GGMR01009076 MBY21695.1 GFDF01005173 JAV08911.1 ABLF02039514 GFDF01005174 JAV08910.1 NWSH01000876 PCG73758.1 AJVK01060758 ABLF02039511 ABLF02039515 ABLF02039538 AK341032 BAH71516.1 GALA01000821 JAA94031.1 GBXI01012537 JAD01755.1 GEZM01006616 JAV95682.1 GFDG01002568 JAV16231.1 GFDG01002571 JAV16228.1 GFDG01002974 JAV15825.1 GFDG01002570 JAV16229.1 GFDG01002569 JAV16230.1 AXCN02002069 GAMC01004142 JAC02414.1 APCN01003375 KX270974 AQT33307.1 JTDY01005653 KOB66777.1 KX270975 AQT33309.1 MH366181 QBB01990.1 GDHF01023526 JAI28788.1 NNAY01000752 OXU26688.1 AAAB01008984 EAA14862.4 GAKP01006331 JAC52621.1 GALX01002873 JAB65593.1 KX270976 AQT33308.1 GAKP01006330 JAC52622.1 GBYB01012698 JAG82465.1 AXCM01000345 RSAL01000212 RVE44175.1 GL764397 EFZ17714.1 GL888048 EGI68836.1 GFXV01007777 MBW19582.1 KQ435710 KOX79734.1 ADTU01018970 UFQT01001398 SSX30377.1 GL438248 EFN69213.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000069272
UP000000673
+ More
UP000008820 UP000030765 UP000069940 UP000249989 UP000002320 UP000007819 UP000095301 UP000075880 UP000030742 UP000019118 UP000192223 UP000095300 UP000218220 UP000092462 UP000075900 UP000075885 UP000075886 UP000075840 UP000037510 UP000215335 UP000075903 UP000075902 UP000007062 UP000075881 UP000076407 UP000076408 UP000075882 UP000075883 UP000002358 UP000075884 UP000007755 UP000053105 UP000005205 UP000075920 UP000000311
UP000008820 UP000030765 UP000069940 UP000249989 UP000002320 UP000007819 UP000095301 UP000075880 UP000030742 UP000019118 UP000192223 UP000095300 UP000218220 UP000092462 UP000075900 UP000075885 UP000075886 UP000075840 UP000037510 UP000215335 UP000075903 UP000075902 UP000007062 UP000075881 UP000076407 UP000076408 UP000075882 UP000075883 UP000002358 UP000075884 UP000007755 UP000053105 UP000005205 UP000075920 UP000000311
Pfam
PF01161 PBP
Interpro
SUPFAM
SSF49777
SSF49777
Gene 3D
CDD
ProteinModelPortal
H9JUQ4
A0A2H1VKW9
A0A2H1X2U3
A0A3S2PKG7
A0A212F259
A0A194PJH0
+ More
I4DNG3 A0A2M3YWB1 A0A2M3Z7V4 A0A182F1E0 W5J4A4 A0A2M4A958 Q1HPY7 Q1HPY8 T1DN25 A0A2M3Z9F2 Q1HQT8 Q16LF6 A0A2M4BZN8 I4DM94 A0A084VL08 S4PMB7 I4DIX6 A0A194PIN5 A0A023EIJ5 A0A023ELU7 A0A0K8TS78 A0A023EJG8 A0A2H8TK93 A0A2S2QI33 B0WRQ3 C4WTE3 U5EQ08 T1P8H0 A0A1I8MNV8 C4WTE2 A0A2H1X2Y7 A0A182IZ08 U4U508 A0A212F257 N6TQS9 A0A1Q3FE74 A0A1L8DR19 A0A1Y1NCM5 A0A1W4X0J4 A0A2W1BIB2 A0A2S2NX04 A0A1L8DR75 J9JYE5 A0A1L8DRE0 A0A1I8Q1V0 A0A2A4JPX5 A0A1W4XB58 A0A1B0DH84 A0A1W4WJY1 C4WUE6 T1DIR4 A0A0A1WTC4 A0A1Y1NCL9 A0A182RSJ7 A0A1L8EBW4 A0A1L8EBV7 A0A1L8EB45 A0A1L8EC88 A0A1L8EBW5 A0A182P8B1 A0A182QV86 W8C068 A0A182I7A8 A0A219YQA9 A0A0L7KU41 A0A219YQ51 A0A411G7T8 A0A0K8UQL9 A0A232F887 A0A182UQ84 A0A182TEW0 Q7PWN1 A0A034WAM2 A0A182JUM3 A0A182XNP9 A0A182Y0B3 V5GYS1 A0A219YR16 A0A034WE87 A0A0C9QE70 A0A182LMV6 A0A182MJ69 K7IVL8 A0A182MZ75 A0A437B228 E9IP20 F4WAF3 K7IVM0 A0A2H8U169 A0A0N0BJX0 A0A158NKV7 A0A182VUD7 A0A336MQ06 E2ABH4
I4DNG3 A0A2M3YWB1 A0A2M3Z7V4 A0A182F1E0 W5J4A4 A0A2M4A958 Q1HPY7 Q1HPY8 T1DN25 A0A2M3Z9F2 Q1HQT8 Q16LF6 A0A2M4BZN8 I4DM94 A0A084VL08 S4PMB7 I4DIX6 A0A194PIN5 A0A023EIJ5 A0A023ELU7 A0A0K8TS78 A0A023EJG8 A0A2H8TK93 A0A2S2QI33 B0WRQ3 C4WTE3 U5EQ08 T1P8H0 A0A1I8MNV8 C4WTE2 A0A2H1X2Y7 A0A182IZ08 U4U508 A0A212F257 N6TQS9 A0A1Q3FE74 A0A1L8DR19 A0A1Y1NCM5 A0A1W4X0J4 A0A2W1BIB2 A0A2S2NX04 A0A1L8DR75 J9JYE5 A0A1L8DRE0 A0A1I8Q1V0 A0A2A4JPX5 A0A1W4XB58 A0A1B0DH84 A0A1W4WJY1 C4WUE6 T1DIR4 A0A0A1WTC4 A0A1Y1NCL9 A0A182RSJ7 A0A1L8EBW4 A0A1L8EBV7 A0A1L8EB45 A0A1L8EC88 A0A1L8EBW5 A0A182P8B1 A0A182QV86 W8C068 A0A182I7A8 A0A219YQA9 A0A0L7KU41 A0A219YQ51 A0A411G7T8 A0A0K8UQL9 A0A232F887 A0A182UQ84 A0A182TEW0 Q7PWN1 A0A034WAM2 A0A182JUM3 A0A182XNP9 A0A182Y0B3 V5GYS1 A0A219YR16 A0A034WE87 A0A0C9QE70 A0A182LMV6 A0A182MJ69 K7IVL8 A0A182MZ75 A0A437B228 E9IP20 F4WAF3 K7IVM0 A0A2H8U169 A0A0N0BJX0 A0A158NKV7 A0A182VUD7 A0A336MQ06 E2ABH4
PDB
5TVD
E-value=1.6246e-52,
Score=516
Ontologies
KEGG
GO
PANTHER
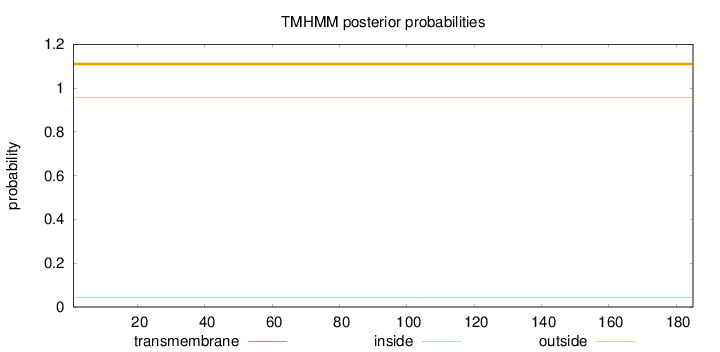
Topology
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0014
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04338
outside
1 - 185
Population Genetic Test Statistics
Pi
246.942003
Theta
156.188245
Tajima's D
2.548037
CLR
0.167744
CSRT
0.948952552372381
Interpretation
Uncertain
