Gene
KWMTBOMO09887
Pre Gene Modal
BGIBMGA013162
Annotation
PREDICTED:_uncharacterized_protein_LOC105388832_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 1.514 Nuclear Reliability : 1.885
Sequence
CDS
ATGACTAAGTTCAGTGGCGAAGATCACACTTACACCTCAACGAAATGGGCGCAGGATCTAGAAGACAACGCCGAAATATTTGGATGGTCGCCTCAACAACTACTTATCATAGCACGTAGATCTTTATCAGGAACAGCACAGTTATGGTTAAAGACGGAGAAGGTTCATAAAAGTTATGAAGAACTCAAGGCAGCCCTGCTCAAAGAATTTCCTGACGTGATGAACAGCAAAGAGATGCATGAGCTCATGACTGCGCGCAAGAAACATAAAGACGAGTCATATTATCAATATATGCTAATTATGAAGGAAATGGGAAAACGAGCGAGATTCCCAGATTACGTTGCCATTCAGTACATCATAGACGGCATTACAGACCTGGAGATGAATAAAGCAATTTTGTACGGTGCGGTTACTTATCCGGTGCTGAAAGAAAAGTTGTTGTTGTACGAAAGGATCAAGGAGAAATCTGCCAAACAGAATGCTGAACTACATATTAAACGAACTGCAACGACTTCGGTTAGAGATTCCATGTCAAGAACCAACCAGTATTGTGCAATAAATTTGGCCATATTAGTTCAAAGTGCATCAATTCTAACACAGGAAGTACCGAAAGTTCGAGTTACACGAAAACGTCGGTAA
Protein
MTKFSGEDHTYTSTKWAQDLEDNAEIFGWSPQQLLIIARRSLSGTAQLWLKTEKVHKSYEELKAALLKEFPDVMNSKEMHELMTARKKHKDESYYQYMLIMKEMGKRARFPDYVAIQYIIDGITDLEMNKAILYGAVTYPVLKEKLLLYERIKEKSAKQNAELHIKRTATTSVRDSMSRTNQYCAINLAILVQSASILTQEVPKVRVTRKRR
Summary
Uniprot
A0A2A4J934
A0A2A4JH77
A0A2A4JEK2
A0A2A4JPY0
A0A2A4J1N2
A0A2A4K4A8
+ More
A0A2A4K0J9 A0A2H1VAI8 A0A2A4J7U9 A0A2A4JBF8 A0A437AU02 T1H9C0 A0A023EYH9 A0A023EZG2 J9HJJ8 A0A0A1XB16 W8BB66 A0A023F013 A0A3L8DTV3 A0A0J7KCU5 A0A139WKB5 A0A0L7QJU0 A0A0K8U1E2 A0A0J7K9X8 A0A0A1X9Z0 J9JZ77 A0A023EY26 A0A139WE33 D7ELA7 A0A034WUA4 A0A1W7R6L8 X1X261 A0A0A1XFE9 A0A034WS55 A0A0K8U2D9 W8B0V9 A0A212FP93 A0A139WB30 W8APB2 W8C3C0 A0A0A9YCQ1 K7JIL1 A0A2M3Z2X0 A0A1S4HEU1 A0A0E4G8D8 A0A084VMA0 W8C8Q1 T1IC96 J9JNX9 A0A1B0CKJ5 A0A1I8NEC0 A0A1I8NEC4 A0A182W804 A0A2M3Z3Z7
A0A2A4K0J9 A0A2H1VAI8 A0A2A4J7U9 A0A2A4JBF8 A0A437AU02 T1H9C0 A0A023EYH9 A0A023EZG2 J9HJJ8 A0A0A1XB16 W8BB66 A0A023F013 A0A3L8DTV3 A0A0J7KCU5 A0A139WKB5 A0A0L7QJU0 A0A0K8U1E2 A0A0J7K9X8 A0A0A1X9Z0 J9JZ77 A0A023EY26 A0A139WE33 D7ELA7 A0A034WUA4 A0A1W7R6L8 X1X261 A0A0A1XFE9 A0A034WS55 A0A0K8U2D9 W8B0V9 A0A212FP93 A0A139WB30 W8APB2 W8C3C0 A0A0A9YCQ1 K7JIL1 A0A2M3Z2X0 A0A1S4HEU1 A0A0E4G8D8 A0A084VMA0 W8C8Q1 T1IC96 J9JNX9 A0A1B0CKJ5 A0A1I8NEC0 A0A1I8NEC4 A0A182W804 A0A2M3Z3Z7
Pubmed
EMBL
NWSH01002403
PCG68369.1
NWSH01001586
PCG70783.1
NWSH01001874
PCG69832.1
+ More
NWSH01000799 PCG74125.1 NWSH01004261 PCG65293.1 NWSH01000198 PCG78490.1 NWSH01000337 PCG77293.1 ODYU01001505 SOQ37806.1 NWSH01002797 PCG67520.1 NWSH01002225 PCG68884.1 RSAL01000438 RVE41711.1 ACPB03028153 GBBI01004550 JAC14162.1 GBBI01004551 JAC14161.1 CH478509 EJY58114.1 GBXI01005773 JAD08519.1 GAMC01016174 JAB90381.1 GBBI01004613 JAC14099.1 QOIP01000004 RLU23736.1 LBMM01009423 KMQ88147.1 KQ971327 KYB28419.1 KQ415113 KOC58816.1 GDHF01031998 JAI20316.1 LBMM01010976 KMQ87102.1 GBXI01006173 JAD08119.1 ABLF02014660 ABLF02042774 GBBI01004477 JAC14235.1 KQ971355 KYB26210.1 KQ972211 EFA11958.2 GAKP01001187 JAC57765.1 GEHC01000930 JAV46715.1 ABLF02005127 ABLF02005129 ABLF02007401 GBXI01004632 JAD09660.1 GAKP01000546 JAC58406.1 GDHF01031400 JAI20914.1 GAMC01015907 JAB90648.1 AGBW02003316 OWR55561.1 KQ971376 KYB25116.1 GAMC01020207 JAB86348.1 GAMC01002792 JAC03764.1 GBHO01014721 JAG28883.1 GGFM01002109 MBW22860.1 AAAB01008984 HACL01000135 CFW94429.1 ATLV01014583 KE524975 KFB39094.1 GAMC01007326 JAB99229.1 ACPB03027146 ABLF02011059 AJWK01016333 GGFM01002486 MBW23237.1
NWSH01000799 PCG74125.1 NWSH01004261 PCG65293.1 NWSH01000198 PCG78490.1 NWSH01000337 PCG77293.1 ODYU01001505 SOQ37806.1 NWSH01002797 PCG67520.1 NWSH01002225 PCG68884.1 RSAL01000438 RVE41711.1 ACPB03028153 GBBI01004550 JAC14162.1 GBBI01004551 JAC14161.1 CH478509 EJY58114.1 GBXI01005773 JAD08519.1 GAMC01016174 JAB90381.1 GBBI01004613 JAC14099.1 QOIP01000004 RLU23736.1 LBMM01009423 KMQ88147.1 KQ971327 KYB28419.1 KQ415113 KOC58816.1 GDHF01031998 JAI20316.1 LBMM01010976 KMQ87102.1 GBXI01006173 JAD08119.1 ABLF02014660 ABLF02042774 GBBI01004477 JAC14235.1 KQ971355 KYB26210.1 KQ972211 EFA11958.2 GAKP01001187 JAC57765.1 GEHC01000930 JAV46715.1 ABLF02005127 ABLF02005129 ABLF02007401 GBXI01004632 JAD09660.1 GAKP01000546 JAC58406.1 GDHF01031400 JAI20914.1 GAMC01015907 JAB90648.1 AGBW02003316 OWR55561.1 KQ971376 KYB25116.1 GAMC01020207 JAB86348.1 GAMC01002792 JAC03764.1 GBHO01014721 JAG28883.1 GGFM01002109 MBW22860.1 AAAB01008984 HACL01000135 CFW94429.1 ATLV01014583 KE524975 KFB39094.1 GAMC01007326 JAB99229.1 ACPB03027146 ABLF02011059 AJWK01016333 GGFM01002486 MBW23237.1
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR000477 RT_dom
IPR018061 Retropepsins
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR032071 DUF4806
IPR041373 RT_RNaseH
IPR003034 SAP_dom
IPR002401 Cyt_P450_E_grp-I
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR036396 Cyt_P450_sf
IPR036361 SAP_dom_sf
IPR005162 Retrotrans_gag_dom
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR019103 Peptidase_aspartic_DDI1-type
IPR021896 Transposase_37
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR000477 RT_dom
IPR018061 Retropepsins
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR032071 DUF4806
IPR041373 RT_RNaseH
IPR003034 SAP_dom
IPR002401 Cyt_P450_E_grp-I
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR036396 Cyt_P450_sf
IPR036361 SAP_dom_sf
IPR005162 Retrotrans_gag_dom
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR019103 Peptidase_aspartic_DDI1-type
IPR021896 Transposase_37
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4J934
A0A2A4JH77
A0A2A4JEK2
A0A2A4JPY0
A0A2A4J1N2
A0A2A4K4A8
+ More
A0A2A4K0J9 A0A2H1VAI8 A0A2A4J7U9 A0A2A4JBF8 A0A437AU02 T1H9C0 A0A023EYH9 A0A023EZG2 J9HJJ8 A0A0A1XB16 W8BB66 A0A023F013 A0A3L8DTV3 A0A0J7KCU5 A0A139WKB5 A0A0L7QJU0 A0A0K8U1E2 A0A0J7K9X8 A0A0A1X9Z0 J9JZ77 A0A023EY26 A0A139WE33 D7ELA7 A0A034WUA4 A0A1W7R6L8 X1X261 A0A0A1XFE9 A0A034WS55 A0A0K8U2D9 W8B0V9 A0A212FP93 A0A139WB30 W8APB2 W8C3C0 A0A0A9YCQ1 K7JIL1 A0A2M3Z2X0 A0A1S4HEU1 A0A0E4G8D8 A0A084VMA0 W8C8Q1 T1IC96 J9JNX9 A0A1B0CKJ5 A0A1I8NEC0 A0A1I8NEC4 A0A182W804 A0A2M3Z3Z7
A0A2A4K0J9 A0A2H1VAI8 A0A2A4J7U9 A0A2A4JBF8 A0A437AU02 T1H9C0 A0A023EYH9 A0A023EZG2 J9HJJ8 A0A0A1XB16 W8BB66 A0A023F013 A0A3L8DTV3 A0A0J7KCU5 A0A139WKB5 A0A0L7QJU0 A0A0K8U1E2 A0A0J7K9X8 A0A0A1X9Z0 J9JZ77 A0A023EY26 A0A139WE33 D7ELA7 A0A034WUA4 A0A1W7R6L8 X1X261 A0A0A1XFE9 A0A034WS55 A0A0K8U2D9 W8B0V9 A0A212FP93 A0A139WB30 W8APB2 W8C3C0 A0A0A9YCQ1 K7JIL1 A0A2M3Z2X0 A0A1S4HEU1 A0A0E4G8D8 A0A084VMA0 W8C8Q1 T1IC96 J9JNX9 A0A1B0CKJ5 A0A1I8NEC0 A0A1I8NEC4 A0A182W804 A0A2M3Z3Z7
Ontologies
KEGG
GO
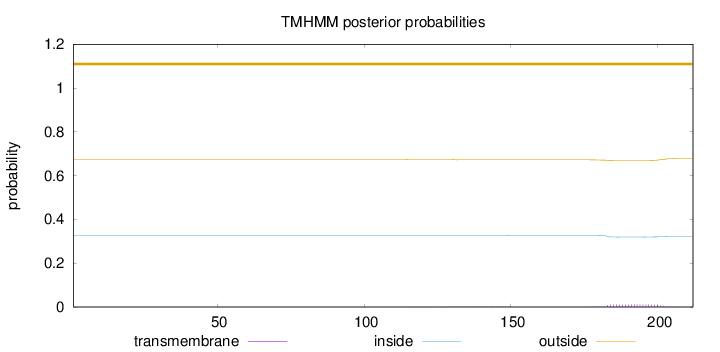
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25982
Exp number, first 60 AAs:
0.00134
Total prob of N-in:
0.32635
outside
1 - 212
Population Genetic Test Statistics
Pi
335.196001
Theta
181.50217
Tajima's D
2.51194
CLR
0.006301
CSRT
0.945152742362882
Interpretation
Uncertain
