Gene
KWMTBOMO09880 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013272
Annotation
PREDICTED:_uncharacterized_protein_LOC101746829_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.559
Sequence
CDS
ATGCCGGCGCGAAAGCGTATTAGTCTGGAGGAAGAGTATAGTAAGCCGACTGGTATCACTGCAGATGATATTGCCAAGCTACGCCAATGGTTGGACACTCAACCGCATCTGCCGAAGAACCTTACCGATCTAGATCTGATCCTGATCTACCACAAATGCGACCGCAGCATCCAAGTCTCCAAACAGGAGATCGATAATCAGTATACCCTAAGAACGTTGTTTACATCCTTCTTCAAGGACCGAAGCTTAAATAAGGATATTGAAAATTTCTTCCTGACCGCGCTCGTCACTATCCTGCCGAATCGCTCCAAAGCGGGCGATGCGATCTTTTACGCTAGGATTTTGGACACAGATGTCAACAAGTTTGACTTCGCCAGCGCTATTAAGGGTGTCTTTATGATGCTGGACCTATGGCAGTACGAGGAAGGAACATGGCCAGGACTGACCTTCATCTTCGATTTGCAAGGAATAAAAATGGGCTTACTGTCCAAATTGGACATACAGAATATACAACAGTTCGTAGCTTATTTCCCTGATGCAATATTAGCGAAATTCAATGGTCTGCACTTCATAAATACGCCCACGTTCATGGACCGCGTGAACACACTCACAAAAGCGTTCTACAAGAATGAGAATATTCAGAAGTTCATCGTCCATCAAAACGGTAGCAACACCTTAGAGCAGATCATTGATATTGATATCTTGCCTAAGGAAGCTGGTGGCAAGTTTAAGAGCTTAGAGGAGTGTCAACAAGATACATTAGAAAAAATACGAGCAAACGAAGATTATTTCGTCGAGGAAAACAAGAAGAGGATCGTTGAAGCACTAAGACCTGGAAAACCGAAGACCATAGCTGATTTCTTTGATAGTGTCGAGGGCTCCTTCAAGAAACTGGAAATTGATTAG
Protein
MPARKRISLEEEYSKPTGITADDIAKLRQWLDTQPHLPKNLTDLDLILIYHKCDRSIQVSKQEIDNQYTLRTLFTSFFKDRSLNKDIENFFLTALVTILPNRSKAGDAIFYARILDTDVNKFDFASAIKGVFMMLDLWQYEEGTWPGLTFIFDLQGIKMGLLSKLDIQNIQQFVAYFPDAILAKFNGLHFINTPTFMDRVNTLTKAFYKNENIQKFIVHQNGSNTLEQIIDIDILPKEAGGKFKSLEECQQDTLEKIRANEDYFVEENKKRIVEALRPGKPKTIADFFDSVEGSFKKLEID
Summary
Uniprot
H9JUQ9
H9JUE6
A0A2H1V703
A0A194PT99
A0A2H4RMT6
A0A2H1VNR9
+ More
A0A0S2Z3U0 A0A2H1VNP8 A0A212F255 A0A2A4JBJ3 A0A2A4IZQ3 A0A437BSJ4 A0A2H4RMR3 A0A212F272 A0A3S2L272 H9JUR0 I4DLH3 A0A212F242 A0A2H4RMP1 A0A0S2Z313 H9IUL0 A0A2H4RMW0 A0A0N0PCB2 A0A2A4JLN7 A0A194PRP9 A0A437B632 H9JUQ5 A0A2A4J9W6 A0A212F251 A0A2H4RMR0 A0A1W4X5F9 A0A2A4ISL2 A0A310SB26 A0A0L7R9P5 V9IMD6 E2A8Q9 A0A088AC26 K7J0H8 A0A2A3EJJ4 A0A2P8XSZ9 E2BL70 A0A1I8PTI0 V9IER4 A0A310SFZ3 A0A151HZ83 A0A1Y1JXE4 A0A158NIC6 F4WLR1 A0A151IIH9 A0A1Y1L4D8 A0A026VZY9 A0A1I8Q2Q3 A0A2A3EI39 K7ITY7 A0A195CB81 A0A151WMV2 A0A0M9A4P5 A0A0S2Z3V6 A0A151WMV5 A0A154PLP4 A0A0L7LLE7 A0A0L7R9L5 A0A139WDA0 A0A0J7NH54 A0A1I8N3L8 A0A1B0FV49 K7J0H5 W8C0A2 A0A2S2Q3J3 A0A195ECZ2 A0A232F895 E9IA70 A0A158NA70 A0A195BKL4 E2A8R3 A0A0L0CK65 A0A151X7H5 A0A151JTT8 A0A195DLY0 A0A195ECX6 A0A1Q3FQZ8 A0A151JTJ8 A0A158NIC8 K7ITY6 A0A067QI19 A0A195FRC0 A0A1B0D1Y1 A0A151WMU4 A0A154PLR6 A0A151HZE1 A0A158NIC7 F4X1H6 A0A151JTJ2 B4G2X4 E2BL75 J9JNK6 Q297R5 A0A0C9RRA5 F4WEQ5
A0A0S2Z3U0 A0A2H1VNP8 A0A212F255 A0A2A4JBJ3 A0A2A4IZQ3 A0A437BSJ4 A0A2H4RMR3 A0A212F272 A0A3S2L272 H9JUR0 I4DLH3 A0A212F242 A0A2H4RMP1 A0A0S2Z313 H9IUL0 A0A2H4RMW0 A0A0N0PCB2 A0A2A4JLN7 A0A194PRP9 A0A437B632 H9JUQ5 A0A2A4J9W6 A0A212F251 A0A2H4RMR0 A0A1W4X5F9 A0A2A4ISL2 A0A310SB26 A0A0L7R9P5 V9IMD6 E2A8Q9 A0A088AC26 K7J0H8 A0A2A3EJJ4 A0A2P8XSZ9 E2BL70 A0A1I8PTI0 V9IER4 A0A310SFZ3 A0A151HZ83 A0A1Y1JXE4 A0A158NIC6 F4WLR1 A0A151IIH9 A0A1Y1L4D8 A0A026VZY9 A0A1I8Q2Q3 A0A2A3EI39 K7ITY7 A0A195CB81 A0A151WMV2 A0A0M9A4P5 A0A0S2Z3V6 A0A151WMV5 A0A154PLP4 A0A0L7LLE7 A0A0L7R9L5 A0A139WDA0 A0A0J7NH54 A0A1I8N3L8 A0A1B0FV49 K7J0H5 W8C0A2 A0A2S2Q3J3 A0A195ECZ2 A0A232F895 E9IA70 A0A158NA70 A0A195BKL4 E2A8R3 A0A0L0CK65 A0A151X7H5 A0A151JTT8 A0A195DLY0 A0A195ECX6 A0A1Q3FQZ8 A0A151JTJ8 A0A158NIC8 K7ITY6 A0A067QI19 A0A195FRC0 A0A1B0D1Y1 A0A151WMU4 A0A154PLR6 A0A151HZE1 A0A158NIC7 F4X1H6 A0A151JTJ2 B4G2X4 E2BL75 J9JNK6 Q297R5 A0A0C9RRA5 F4WEQ5
Pubmed
EMBL
BABH01022835
BABH01022836
BABH01022838
ODYU01000988
SOQ36579.1
KQ459595
+ More
KPI95989.1 MG434644 ATY51950.1 ODYU01003540 SOQ42448.1 KT943550 ALQ33308.1 ODYU01003541 SOQ42450.1 AGBW02010783 OWR47825.1 NWSH01002010 PCG69445.1 NWSH01004847 PCG64660.1 RSAL01000013 RVE53444.1 MG434617 ATY51923.1 OWR47826.1 RSAL01000223 RVE44001.1 BABH01022842 BABH01022843 BABH01022844 AK402141 BAM18763.1 KPI95986.1 OWR47818.1 MG434601 ATY51907.1 KT943538 ALQ33296.1 BABH01000888 MG434659 ATY51965.1 KQ460643 KPJ13319.1 NWSH01001157 PCG72320.1 KPI95987.1 RSAL01000144 RVE45960.1 BABH01022824 NWSH01002262 PCG68761.1 OWR47817.1 MG434609 ATY51915.1 NWSH01007421 PCG62977.1 KQ774368 OAD52321.1 KQ414624 KOC67554.1 JR052704 AEY61761.1 GL437663 EFN70138.1 KZ288238 PBC31369.1 PYGN01001396 PSN35126.1 GL448949 EFN83556.1 JR042329 AEY59598.1 OAD52320.1 KQ976695 KYM77582.1 GEZM01098136 JAV53982.1 ADTU01016468 GL888213 EGI64859.1 KQ977477 KYN02476.1 GEZM01068032 JAV67250.1 KK107536 EZA49210.1 PBC31368.1 KQ978009 KYM98124.1 KQ982934 KYQ49138.1 KQ435735 KOX77180.1 KT943566 ALQ33324.1 KYQ49137.1 KQ434973 KZC12772.1 JTDY01000701 KOB76164.1 KOC67553.1 KQ971361 KYB25884.1 LBMM01005052 KMQ91855.1 AJWK01010375 GAMC01011526 JAB95029.1 GGMS01003091 MBY72294.1 KQ979074 KYN23073.1 NNAY01000747 OXU26710.1 GL761987 EFZ22528.1 ADTU01010052 KQ976455 KYM85150.1 EFN70142.1 JRES01000266 KNC32793.1 KQ982446 KYQ56322.1 KQ981875 KYN34011.1 KQ980734 KYN13837.1 KYN23070.1 GFDL01005015 JAV30030.1 KYN34015.1 ADTU01016481 ADTU01016482 ADTU01016483 ADTU01016484 KK853366 KDR08117.1 KQ981305 KYN42976.1 AJVK01022454 AJVK01022455 AJVK01022456 KYQ49140.1 KZC12773.1 KYM77581.1 ADTU01016476 ADTU01016477 GL888529 EGI59704.1 KYN34006.1 CH479179 EDW24169.1 EFN83561.1 ABLF02013760 CM000070 EAL28140.1 GBYB01009841 JAG79608.1 GL888105 EGI67317.1
KPI95989.1 MG434644 ATY51950.1 ODYU01003540 SOQ42448.1 KT943550 ALQ33308.1 ODYU01003541 SOQ42450.1 AGBW02010783 OWR47825.1 NWSH01002010 PCG69445.1 NWSH01004847 PCG64660.1 RSAL01000013 RVE53444.1 MG434617 ATY51923.1 OWR47826.1 RSAL01000223 RVE44001.1 BABH01022842 BABH01022843 BABH01022844 AK402141 BAM18763.1 KPI95986.1 OWR47818.1 MG434601 ATY51907.1 KT943538 ALQ33296.1 BABH01000888 MG434659 ATY51965.1 KQ460643 KPJ13319.1 NWSH01001157 PCG72320.1 KPI95987.1 RSAL01000144 RVE45960.1 BABH01022824 NWSH01002262 PCG68761.1 OWR47817.1 MG434609 ATY51915.1 NWSH01007421 PCG62977.1 KQ774368 OAD52321.1 KQ414624 KOC67554.1 JR052704 AEY61761.1 GL437663 EFN70138.1 KZ288238 PBC31369.1 PYGN01001396 PSN35126.1 GL448949 EFN83556.1 JR042329 AEY59598.1 OAD52320.1 KQ976695 KYM77582.1 GEZM01098136 JAV53982.1 ADTU01016468 GL888213 EGI64859.1 KQ977477 KYN02476.1 GEZM01068032 JAV67250.1 KK107536 EZA49210.1 PBC31368.1 KQ978009 KYM98124.1 KQ982934 KYQ49138.1 KQ435735 KOX77180.1 KT943566 ALQ33324.1 KYQ49137.1 KQ434973 KZC12772.1 JTDY01000701 KOB76164.1 KOC67553.1 KQ971361 KYB25884.1 LBMM01005052 KMQ91855.1 AJWK01010375 GAMC01011526 JAB95029.1 GGMS01003091 MBY72294.1 KQ979074 KYN23073.1 NNAY01000747 OXU26710.1 GL761987 EFZ22528.1 ADTU01010052 KQ976455 KYM85150.1 EFN70142.1 JRES01000266 KNC32793.1 KQ982446 KYQ56322.1 KQ981875 KYN34011.1 KQ980734 KYN13837.1 KYN23070.1 GFDL01005015 JAV30030.1 KYN34015.1 ADTU01016481 ADTU01016482 ADTU01016483 ADTU01016484 KK853366 KDR08117.1 KQ981305 KYN42976.1 AJVK01022454 AJVK01022455 AJVK01022456 KYQ49140.1 KZC12773.1 KYM77581.1 ADTU01016476 ADTU01016477 GL888529 EGI59704.1 KYN34006.1 CH479179 EDW24169.1 EFN83561.1 ABLF02013760 CM000070 EAL28140.1 GBYB01009841 JAG79608.1 GL888105 EGI67317.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000283053
UP000053240
+ More
UP000192223 UP000053825 UP000000311 UP000005203 UP000002358 UP000242457 UP000245037 UP000008237 UP000095300 UP000078540 UP000005205 UP000007755 UP000078542 UP000053097 UP000075809 UP000053105 UP000076502 UP000037510 UP000007266 UP000036403 UP000095301 UP000092461 UP000078492 UP000215335 UP000037069 UP000078541 UP000027135 UP000092462 UP000008744 UP000007819 UP000001819
UP000192223 UP000053825 UP000000311 UP000005203 UP000002358 UP000242457 UP000245037 UP000008237 UP000095300 UP000078540 UP000005205 UP000007755 UP000078542 UP000053097 UP000075809 UP000053105 UP000076502 UP000037510 UP000007266 UP000036403 UP000095301 UP000092461 UP000078492 UP000215335 UP000037069 UP000078541 UP000027135 UP000092462 UP000008744 UP000007819 UP000001819
Interpro
Gene 3D
ProteinModelPortal
H9JUQ9
H9JUE6
A0A2H1V703
A0A194PT99
A0A2H4RMT6
A0A2H1VNR9
+ More
A0A0S2Z3U0 A0A2H1VNP8 A0A212F255 A0A2A4JBJ3 A0A2A4IZQ3 A0A437BSJ4 A0A2H4RMR3 A0A212F272 A0A3S2L272 H9JUR0 I4DLH3 A0A212F242 A0A2H4RMP1 A0A0S2Z313 H9IUL0 A0A2H4RMW0 A0A0N0PCB2 A0A2A4JLN7 A0A194PRP9 A0A437B632 H9JUQ5 A0A2A4J9W6 A0A212F251 A0A2H4RMR0 A0A1W4X5F9 A0A2A4ISL2 A0A310SB26 A0A0L7R9P5 V9IMD6 E2A8Q9 A0A088AC26 K7J0H8 A0A2A3EJJ4 A0A2P8XSZ9 E2BL70 A0A1I8PTI0 V9IER4 A0A310SFZ3 A0A151HZ83 A0A1Y1JXE4 A0A158NIC6 F4WLR1 A0A151IIH9 A0A1Y1L4D8 A0A026VZY9 A0A1I8Q2Q3 A0A2A3EI39 K7ITY7 A0A195CB81 A0A151WMV2 A0A0M9A4P5 A0A0S2Z3V6 A0A151WMV5 A0A154PLP4 A0A0L7LLE7 A0A0L7R9L5 A0A139WDA0 A0A0J7NH54 A0A1I8N3L8 A0A1B0FV49 K7J0H5 W8C0A2 A0A2S2Q3J3 A0A195ECZ2 A0A232F895 E9IA70 A0A158NA70 A0A195BKL4 E2A8R3 A0A0L0CK65 A0A151X7H5 A0A151JTT8 A0A195DLY0 A0A195ECX6 A0A1Q3FQZ8 A0A151JTJ8 A0A158NIC8 K7ITY6 A0A067QI19 A0A195FRC0 A0A1B0D1Y1 A0A151WMU4 A0A154PLR6 A0A151HZE1 A0A158NIC7 F4X1H6 A0A151JTJ2 B4G2X4 E2BL75 J9JNK6 Q297R5 A0A0C9RRA5 F4WEQ5
A0A0S2Z3U0 A0A2H1VNP8 A0A212F255 A0A2A4JBJ3 A0A2A4IZQ3 A0A437BSJ4 A0A2H4RMR3 A0A212F272 A0A3S2L272 H9JUR0 I4DLH3 A0A212F242 A0A2H4RMP1 A0A0S2Z313 H9IUL0 A0A2H4RMW0 A0A0N0PCB2 A0A2A4JLN7 A0A194PRP9 A0A437B632 H9JUQ5 A0A2A4J9W6 A0A212F251 A0A2H4RMR0 A0A1W4X5F9 A0A2A4ISL2 A0A310SB26 A0A0L7R9P5 V9IMD6 E2A8Q9 A0A088AC26 K7J0H8 A0A2A3EJJ4 A0A2P8XSZ9 E2BL70 A0A1I8PTI0 V9IER4 A0A310SFZ3 A0A151HZ83 A0A1Y1JXE4 A0A158NIC6 F4WLR1 A0A151IIH9 A0A1Y1L4D8 A0A026VZY9 A0A1I8Q2Q3 A0A2A3EI39 K7ITY7 A0A195CB81 A0A151WMV2 A0A0M9A4P5 A0A0S2Z3V6 A0A151WMV5 A0A154PLP4 A0A0L7LLE7 A0A0L7R9L5 A0A139WDA0 A0A0J7NH54 A0A1I8N3L8 A0A1B0FV49 K7J0H5 W8C0A2 A0A2S2Q3J3 A0A195ECZ2 A0A232F895 E9IA70 A0A158NA70 A0A195BKL4 E2A8R3 A0A0L0CK65 A0A151X7H5 A0A151JTT8 A0A195DLY0 A0A195ECX6 A0A1Q3FQZ8 A0A151JTJ8 A0A158NIC8 K7ITY6 A0A067QI19 A0A195FRC0 A0A1B0D1Y1 A0A151WMU4 A0A154PLR6 A0A151HZE1 A0A158NIC7 F4X1H6 A0A151JTJ2 B4G2X4 E2BL75 J9JNK6 Q297R5 A0A0C9RRA5 F4WEQ5
PDB
3W68
E-value=4.48519e-09,
Score=145
Ontologies
Topology
Subcellular location
Nucleus
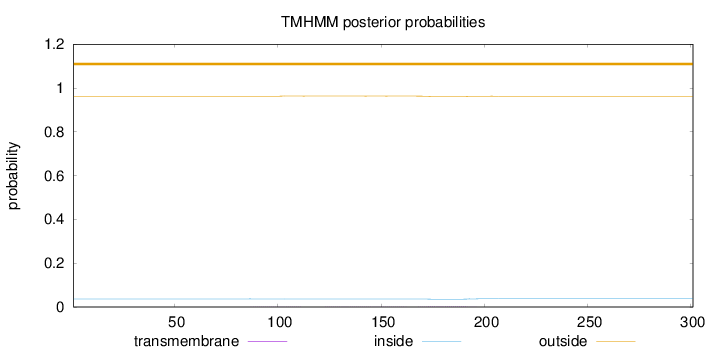
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06308
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03694
outside
1 - 301
Population Genetic Test Statistics
Pi
309.400655
Theta
214.553307
Tajima's D
1.503226
CLR
0
CSRT
0.790810459477026
Interpretation
Uncertain
