Gene
KWMTBOMO09871 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013275
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_carboxypeptidase_B-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.055 Extracellular Reliability : 1.109
Sequence
CDS
ATGGATGCGAGTTTTTTTGTTATTCTCGCTTTAATAAGCTCCGTGTTCGCCGGTAAACACGATATTTACTCCAGACATGCTGTCTACGGTGTTGAGCTTCGAGACCAGGATGATGTGGCGACTCTCTTCAATCTCCAGCAAAAACTGAACGTCGATATCTGGCAGCATGGTATGGCCAGAGTCAGAGACGCTGAAGTCATGGTTTCACCGGAACAGTCCAAGGAGTTCTTCGAGACTTTAGATAATAACGGCTTGAAGTATTATTTGAAAATAGAAGATGTTTCTAAAGTTCTGAAGGAGCACGAAGAGGATCTGGCGAAATGGAGGAGAAACAGAAGTAACAGAATGATCTTTCAAGATTATCCGCGTTACGCTGAGGTAAACCAATACATGGAAGAAATTGCCGCTAGATATCCGAATTTGGTGACCCTAGTCAACGCTGGGAAAAGCTTCGAGGGACGCGACATCAAATATTTGAAGATATCGACAACAAACTTCGAGGATCCCCGCAAACCAGTATACTTTATGGACGCCATGATGCACGCTCGTGAATGGGTAACGACACCAGTGACGATATACTCAGTCTTCAGACTCGTAGAGAACTTGAGAGATGATGAGAGGGATCTGCTTGAAGACATCGATTGGATCATTTTGCCTATTGTCAATCCCGATGGCTATGAGTTCAGTCACACTGACGATCGTCTTTGGCGTCGCACCCGTTCCGTCAACCTAGAAGTCAGTACTACCTGCTTTGGAGTCGACCCGAATAGGAACTTTGACGTCGACTTCAATACCTTAGGGGTCTCCAGCGACCCGTGCTCCCAAACATATCCAGGGGTAGCGCCGTTCTCCGAAGCAGAAACGAGGATTATCAGGGATATCTTCTTGAAGTACTTGCCGCGTATACAGCTGTACAATGATATTCATAGCCATGGAAACTATGTTCTGTTCGGTTTTGGAAACAACACTTTGACGCCTAATGTGGCACAGCTGCATCATGTGGCTGCCGCGATGGGCGCGGCCATGGACGCTGTCAAATTACCCGAAGCCGGATATTATCTGATCGGTAACAGCGGTCTGGTGCTATACAAAACTTCTGGATGCGGCCAGGATTACGGTCAGGTGAGAGCATACATTAGATAA
Protein
MDASFFVILALISSVFAGKHDIYSRHAVYGVELRDQDDVATLFNLQQKLNVDIWQHGMARVRDAEVMVSPEQSKEFFETLDNNGLKYYLKIEDVSKVLKEHEEDLAKWRRNRSNRMIFQDYPRYAEVNQYMEEIAARYPNLVTLVNAGKSFEGRDIKYLKISTTNFEDPRKPVYFMDAMMHAREWVTTPVTIYSVFRLVENLRDDERDLLEDIDWIILPIVNPDGYEFSHTDDRLWRRTRSVNLEVSTTCFGVDPNRNFDVDFNTLGVSSDPCSQTYPGVAPFSEAETRIIRDIFLKYLPRIQLYNDIHSHGNYVLFGFGNNTLTPNVAQLHHVAAAMGAAMDAVKLPEAGYYLIGNSGLVLYKTSGCGQDYGQVRAYIR
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JUR2
H9JUR3
A0A2H1V2Z0
A0A0N1INA8
A0A194PTA4
A0A1E1WRL9
+ More
A0A0N1I4Q3 A0A212FM32 A0A1E1WL84 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 B1NLE5 A0A2A4J4T7 A0A194Q4D5 Q6H961 Q699Z2 A0A194PY54 O97389 O97434 H9JIY5 D2Y4U3 H9JIX6 A0A2H1WC94 A0A437AY42 A0A2A4JD50 A0A194PZ08 A0A212FCH2 H9JIY6 Q699Z1 A0A437AU59 H9J5P0 Q3T905 U3LUX2 H9JIX7 A0A2H1WCA6 A0A0N1PGB0 A0A2H1VYG5 A0A194PM74 Q9TYH8 Q6H962 A0A0N0PC47 A0A3S2P6N8 C9W8K4 A0A194RQH5 A0A2H1W3T9 A0A212F2Y7 A0A2A4J5Z1 B5TGP9 A0A1E1WQE4 Q699Y8 A0A2H1VQN5 Q699Y9 A0A212FCH8 A0A2H1WNJ7 V5G5X5 A0A1B0C648 A0A1A9XE82 J3JW26 A0A0L7KXF0 A0A1I8M460 A0A0L0CSA8 A0A1S4F020 D3TN46 Q6J661 Q17K11 B4HV67 B4QKE7 Q961J8 A0A023ESX1 A0A182GUN0 A0A1V1FQK4 A0A212FCG9 A0A3B0J9V9 A0A1B0GH60 G9B641 A0A336MC79 Q2M134 A0A1W4XSM1 B4ML78 A0A336LV12 Q6J660 B3NFD2 A0A1W4UCC4 A0A1A9VKY8 A0A1W4XH91 B4PK76 A0A1L8DR84 G9B628 G9B639 G9B630 G9B629 G9B645 G9B626 A0A1L8DPE5
A0A0N1I4Q3 A0A212FM32 A0A1E1WL84 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 B1NLE5 A0A2A4J4T7 A0A194Q4D5 Q6H961 Q699Z2 A0A194PY54 O97389 O97434 H9JIY5 D2Y4U3 H9JIX6 A0A2H1WC94 A0A437AY42 A0A2A4JD50 A0A194PZ08 A0A212FCH2 H9JIY6 Q699Z1 A0A437AU59 H9J5P0 Q3T905 U3LUX2 H9JIX7 A0A2H1WCA6 A0A0N1PGB0 A0A2H1VYG5 A0A194PM74 Q9TYH8 Q6H962 A0A0N0PC47 A0A3S2P6N8 C9W8K4 A0A194RQH5 A0A2H1W3T9 A0A212F2Y7 A0A2A4J5Z1 B5TGP9 A0A1E1WQE4 Q699Y8 A0A2H1VQN5 Q699Y9 A0A212FCH8 A0A2H1WNJ7 V5G5X5 A0A1B0C648 A0A1A9XE82 J3JW26 A0A0L7KXF0 A0A1I8M460 A0A0L0CSA8 A0A1S4F020 D3TN46 Q6J661 Q17K11 B4HV67 B4QKE7 Q961J8 A0A023ESX1 A0A182GUN0 A0A1V1FQK4 A0A212FCG9 A0A3B0J9V9 A0A1B0GH60 G9B641 A0A336MC79 Q2M134 A0A1W4XSM1 B4ML78 A0A336LV12 Q6J660 B3NFD2 A0A1W4UCC4 A0A1A9VKY8 A0A1W4XH91 B4PK76 A0A1L8DR84 G9B628 G9B639 G9B630 G9B629 G9B645 G9B626 A0A1L8DPE5
Pubmed
19121390
26354079
22118469
18314941
15128309
15262287
+ More
9404008 9807221 11676544 16260742 20824821 22516182 23537049 26227816 25315136 26108605 20353571 17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 28410430 15632085 17550304
9404008 9807221 11676544 16260742 20824821 22516182 23537049 26227816 25315136 26108605 20353571 17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 28410430 15632085 17550304
EMBL
BABH01022852
BABH01022853
BABH01022854
BABH01022855
BABH01022856
ODYU01000433
+ More
SOQ35191.1 KQ461189 KPJ07296.1 KQ459595 KPI95994.1 GDQN01001497 JAT89557.1 KPJ07295.1 AGBW02007655 OWR54814.1 GDQN01003319 JAT87735.1 GDQN01005910 JAT85144.1 KQ460778 KPJ12425.1 GU325786 ADB96010.1 EF600060 ABU98625.1 NWSH01002977 PCG67157.1 KQ459585 KPI98270.1 AJ626863 CAF25190.1 AY512674 AAS82583.1 KPI98271.1 AJ005177 CAA06418.1 AJ005178 CAA06419.1 BABH01022994 GU323795 ADA83700.1 BABH01022992 ODYU01007689 SOQ50701.1 RSAL01000278 RVE43076.1 NWSH01002042 PCG69323.1 KPI98273.1 AGBW02009192 OWR51432.1 BABH01022995 AY512675 AAS82584.1 RSAL01000517 RVE41479.1 BABH01024959 BABH01024960 AM085495 CAJ30028.1 JX865781 AGL42356.1 BABH01022991 SOQ50698.1 KPJ12423.1 ODYU01005225 SOQ45875.1 KQ459604 KPI92215.1 AJ005176 CAA06417.1 AJ626862 CAF25189.2 KPJ12422.1 RVE43075.1 FJ210817 ACN69214.1 KQ460045 KPJ18276.1 ODYU01006127 SOQ47708.1 AGBW02010665 OWR48083.1 PCG67156.1 EU924507 ACH82086.1 GDQN01002013 JAT89041.1 AY512678 AAS82587.1 ODYU01003862 SOQ43155.1 AY512677 AAS82586.1 OWR51430.1 ODYU01009870 SOQ54577.1 GALX01003006 JAB65460.1 JXJN01026429 BT127444 KB631870 AEE62406.1 ERL86847.1 JTDY01004612 KOB67948.1 JRES01000080 KNC34299.1 CCAG010004976 EZ422848 ADD19124.1 AY590494 AY593982 AAT36735.1 CH477229 EAT46987.1 CH480817 EDW50838.1 CM000363 CM002912 EDX09570.1 KMY98094.1 AE014296 AY051552 AAF50560.2 AAK92976.1 GAPW01001697 JAC11901.1 JXUM01000712 FX985400 BAX07413.1 OWR51431.1 OUUW01000002 SPP77183.1 AJWK01003990 HM543508 AEI58659.1 UFQS01000637 UFQT01000637 SSX05638.1 SSX25997.1 CH379069 EAL30738.1 CH963847 EDW73136.1 UFQT01000127 SSX20529.1 AY593983 AAT36736.1 CH954178 EDV50474.1 CM000159 EDW93756.1 GFDF01005234 JAV08850.1 HM543495 AEI58646.1 HM543506 AEI58657.1 HM543497 AEI58648.1 HM543496 AEI58647.1 HM543512 AEI58663.1 HM543493 AEI58644.1 GFDF01005765 JAV08319.1
SOQ35191.1 KQ461189 KPJ07296.1 KQ459595 KPI95994.1 GDQN01001497 JAT89557.1 KPJ07295.1 AGBW02007655 OWR54814.1 GDQN01003319 JAT87735.1 GDQN01005910 JAT85144.1 KQ460778 KPJ12425.1 GU325786 ADB96010.1 EF600060 ABU98625.1 NWSH01002977 PCG67157.1 KQ459585 KPI98270.1 AJ626863 CAF25190.1 AY512674 AAS82583.1 KPI98271.1 AJ005177 CAA06418.1 AJ005178 CAA06419.1 BABH01022994 GU323795 ADA83700.1 BABH01022992 ODYU01007689 SOQ50701.1 RSAL01000278 RVE43076.1 NWSH01002042 PCG69323.1 KPI98273.1 AGBW02009192 OWR51432.1 BABH01022995 AY512675 AAS82584.1 RSAL01000517 RVE41479.1 BABH01024959 BABH01024960 AM085495 CAJ30028.1 JX865781 AGL42356.1 BABH01022991 SOQ50698.1 KPJ12423.1 ODYU01005225 SOQ45875.1 KQ459604 KPI92215.1 AJ005176 CAA06417.1 AJ626862 CAF25189.2 KPJ12422.1 RVE43075.1 FJ210817 ACN69214.1 KQ460045 KPJ18276.1 ODYU01006127 SOQ47708.1 AGBW02010665 OWR48083.1 PCG67156.1 EU924507 ACH82086.1 GDQN01002013 JAT89041.1 AY512678 AAS82587.1 ODYU01003862 SOQ43155.1 AY512677 AAS82586.1 OWR51430.1 ODYU01009870 SOQ54577.1 GALX01003006 JAB65460.1 JXJN01026429 BT127444 KB631870 AEE62406.1 ERL86847.1 JTDY01004612 KOB67948.1 JRES01000080 KNC34299.1 CCAG010004976 EZ422848 ADD19124.1 AY590494 AY593982 AAT36735.1 CH477229 EAT46987.1 CH480817 EDW50838.1 CM000363 CM002912 EDX09570.1 KMY98094.1 AE014296 AY051552 AAF50560.2 AAK92976.1 GAPW01001697 JAC11901.1 JXUM01000712 FX985400 BAX07413.1 OWR51431.1 OUUW01000002 SPP77183.1 AJWK01003990 HM543508 AEI58659.1 UFQS01000637 UFQT01000637 SSX05638.1 SSX25997.1 CH379069 EAL30738.1 CH963847 EDW73136.1 UFQT01000127 SSX20529.1 AY593983 AAT36736.1 CH954178 EDV50474.1 CM000159 EDW93756.1 GFDF01005234 JAV08850.1 HM543495 AEI58646.1 HM543506 AEI58657.1 HM543497 AEI58648.1 HM543496 AEI58647.1 HM543512 AEI58663.1 HM543493 AEI58644.1 GFDF01005765 JAV08319.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000092460 UP000092443 UP000030742 UP000037510 UP000095301 UP000037069 UP000092444 UP000008820 UP000001292 UP000000304 UP000000803 UP000069940 UP000268350 UP000092461 UP000001819 UP000192223 UP000007798 UP000008711 UP000192221 UP000078200 UP000002282
UP000092460 UP000092443 UP000030742 UP000037510 UP000095301 UP000037069 UP000092444 UP000008820 UP000001292 UP000000304 UP000000803 UP000069940 UP000268350 UP000092461 UP000001819 UP000192223 UP000007798 UP000008711 UP000192221 UP000078200 UP000002282
Interpro
Gene 3D
ProteinModelPortal
H9JUR2
H9JUR3
A0A2H1V2Z0
A0A0N1INA8
A0A194PTA4
A0A1E1WRL9
+ More
A0A0N1I4Q3 A0A212FM32 A0A1E1WL84 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 B1NLE5 A0A2A4J4T7 A0A194Q4D5 Q6H961 Q699Z2 A0A194PY54 O97389 O97434 H9JIY5 D2Y4U3 H9JIX6 A0A2H1WC94 A0A437AY42 A0A2A4JD50 A0A194PZ08 A0A212FCH2 H9JIY6 Q699Z1 A0A437AU59 H9J5P0 Q3T905 U3LUX2 H9JIX7 A0A2H1WCA6 A0A0N1PGB0 A0A2H1VYG5 A0A194PM74 Q9TYH8 Q6H962 A0A0N0PC47 A0A3S2P6N8 C9W8K4 A0A194RQH5 A0A2H1W3T9 A0A212F2Y7 A0A2A4J5Z1 B5TGP9 A0A1E1WQE4 Q699Y8 A0A2H1VQN5 Q699Y9 A0A212FCH8 A0A2H1WNJ7 V5G5X5 A0A1B0C648 A0A1A9XE82 J3JW26 A0A0L7KXF0 A0A1I8M460 A0A0L0CSA8 A0A1S4F020 D3TN46 Q6J661 Q17K11 B4HV67 B4QKE7 Q961J8 A0A023ESX1 A0A182GUN0 A0A1V1FQK4 A0A212FCG9 A0A3B0J9V9 A0A1B0GH60 G9B641 A0A336MC79 Q2M134 A0A1W4XSM1 B4ML78 A0A336LV12 Q6J660 B3NFD2 A0A1W4UCC4 A0A1A9VKY8 A0A1W4XH91 B4PK76 A0A1L8DR84 G9B628 G9B639 G9B630 G9B629 G9B645 G9B626 A0A1L8DPE5
A0A0N1I4Q3 A0A212FM32 A0A1E1WL84 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 B1NLE5 A0A2A4J4T7 A0A194Q4D5 Q6H961 Q699Z2 A0A194PY54 O97389 O97434 H9JIY5 D2Y4U3 H9JIX6 A0A2H1WC94 A0A437AY42 A0A2A4JD50 A0A194PZ08 A0A212FCH2 H9JIY6 Q699Z1 A0A437AU59 H9J5P0 Q3T905 U3LUX2 H9JIX7 A0A2H1WCA6 A0A0N1PGB0 A0A2H1VYG5 A0A194PM74 Q9TYH8 Q6H962 A0A0N0PC47 A0A3S2P6N8 C9W8K4 A0A194RQH5 A0A2H1W3T9 A0A212F2Y7 A0A2A4J5Z1 B5TGP9 A0A1E1WQE4 Q699Y8 A0A2H1VQN5 Q699Y9 A0A212FCH8 A0A2H1WNJ7 V5G5X5 A0A1B0C648 A0A1A9XE82 J3JW26 A0A0L7KXF0 A0A1I8M460 A0A0L0CSA8 A0A1S4F020 D3TN46 Q6J661 Q17K11 B4HV67 B4QKE7 Q961J8 A0A023ESX1 A0A182GUN0 A0A1V1FQK4 A0A212FCG9 A0A3B0J9V9 A0A1B0GH60 G9B641 A0A336MC79 Q2M134 A0A1W4XSM1 B4ML78 A0A336LV12 Q6J660 B3NFD2 A0A1W4UCC4 A0A1A9VKY8 A0A1W4XH91 B4PK76 A0A1L8DR84 G9B628 G9B639 G9B630 G9B629 G9B645 G9B626 A0A1L8DPE5
PDB
1JQG
E-value=2.35007e-67,
Score=649
Ontologies
GO
Topology
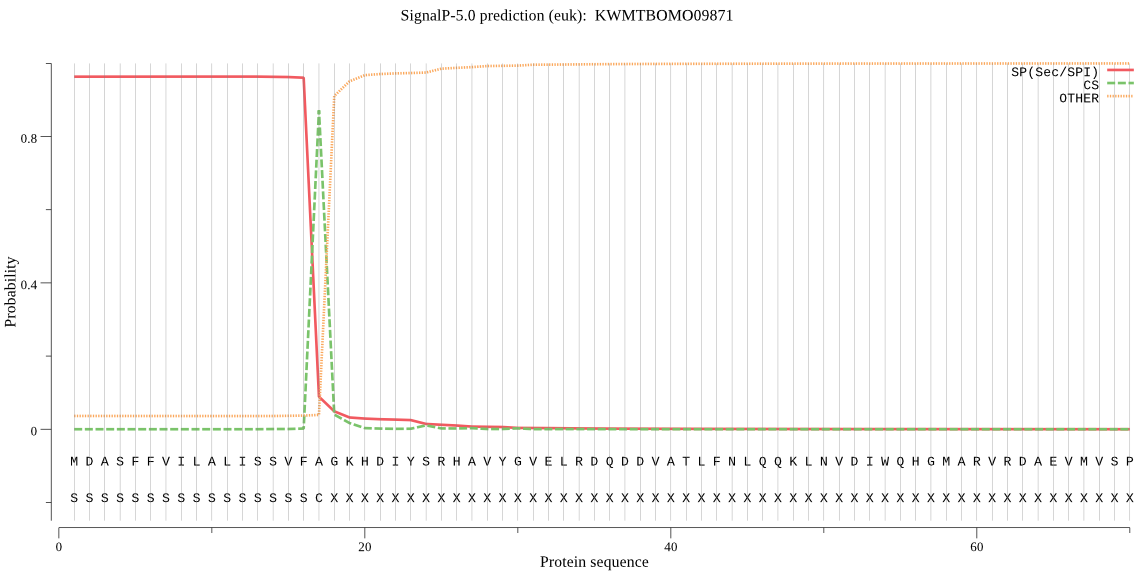
SignalP
Position: 1 - 17,
Likelihood: 0.963013
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43345
Exp number, first 60 AAs:
0.37712
Total prob of N-in:
0.02029
outside
1 - 380

Population Genetic Test Statistics
Pi
290.429803
Theta
200.843928
Tajima's D
1.486643
CLR
0.048591
CSRT
0.781110944452777
Interpretation
Uncertain
