Pre Gene Modal
BGIBMGA013277
Annotation
Uncharacterized_protein_OBRU01_13232_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.463
Sequence
CDS
ATGTCACACACTGTTTTAAGAGGGAAGCCAAACGCTTCTCAACTGTATCTAGATGAAGTGTTAAATGAAATATTAACGGCCAAAGGTGTCGTTCCAGATGCAAAAGAGCGGGATAGATTGATATTAGCAGAAATTGCTTTTCACTGTCTAAAGAAACGACCAAACGCCACAAACATGATAAACGGATCGACTGGAGAAAGTTTCACAAATGAGCAAATTCTGAAACGCGCTGTATCTATTGCACGTTCTATAATGGCCAGAGGGGCCGCAGGGAACAACATTATGGTGGTGATGAGAAACCATCAAAATTTATTTTCTATCTATTGGTCACTACTACTTAGTGGTGCTTTACCTTTCATGATGGATCCCAGCACTACTGTTTATGAACTCGGTTATTTTCTTCAATTACTCGAACCAAGTATAGTGTTTTGTGACCGAGAATATTACAACGACATAAAAAAATCTTTAGATGACTTGCCTGATCTTAAAACAGAGGTTTACATTTGCAACGAAGACGACCTTCTAGAGGATTTTATAAATGGGCATAGTAACGACATCGACAGTTTTAGGATTCCCGAAGGGAATCCAGAGGATACAATATTATTACTACCCACAAGTGGATCCACTGGACTACCTAAAGCCGTACTCTTAACAAATAGGGGAATAGTTGCTCATCTACCTACGTCTTGGACGTATCACACGAAGTTTCCTACGCCAACTGATCTGGTGATGGTATTGACGACAGCGCAATGGATGACGTTTACCATGAAAATCACGACATGCGCCGTTTACCACATACCAATTCTAGTAACTCCCAAGAAAAGAACAGTCGAACATGTCATTGAGATAATCGAGAACTTCCGGCCGACTTGGACGTTTTTCAATCCAGCTTTTGCGAAAGGCTTACTGGCGGTCGTCAGGCGCGAACAGATAGGATCTTTGGAGACCATAATACTCCTCGGATCTCTTGTCACTCCTGATTTAGTTAAGGAATTCAAGGAGAAACTACACCCAGATACACATTTATGTAATGGATATGGGACCACCGAGACACAAGGATTCATCGCTGTGACCGACAGAGACGCGGAGGCTAGCTCGAATGGATGGGTGTATAATATTTTACATTACAAGTTGATCGATGATAATGGCGCTGAAGTGGGTCCAGGGAGTAATGGTGAACTTTACGTTAAAGGCGAATGTGTCGTTAAGGGCTATTACAAAAATAAAGAAGCCTACAGCGAATCCTTTGTGGACGGTTGGTACAAAACGGGAGACTGGTTCCATTTGGATGAGAACGAACGCCTTCACTTTTTGGAGAGGAGGAAATTCAATTTCAAATTCCGGGGGTGTCACGTGTCTCCGGAAGAAGTTGAAGGCGTCATCGGTAAATTACCAGGAGTACACGAGAGCGTGGTGGTGGCAACTGAAATCGGACCCGCTGCAGCTGTAGTCCTCTTACCAGAATACGACCTTACTAGAGAAGATATTAATAAGGCTGTAGACTCAACACTAAGCGATCACAAGAGATTGCACGGAGGCATTGCGTTCGTGGACTCATTGCCGCATACGCACTCAGGAAAACTAAAGAGAATGGAATGCAAGAAGCTCATAGAAGAACTCTTAAAGGATGGAAAATGTTTTTGA
Protein
MSHTVLRGKPNASQLYLDEVLNEILTAKGVVPDAKERDRLILAEIAFHCLKKRPNATNMINGSTGESFTNEQILKRAVSIARSIMARGAAGNNIMVVMRNHQNLFSIYWSLLLSGALPFMMDPSTTVYELGYFLQLLEPSIVFCDREYYNDIKKSLDDLPDLKTEVYICNEDDLLEDFINGHSNDIDSFRIPEGNPEDTILLLPTSGSTGLPKAVLLTNRGIVAHLPTSWTYHTKFPTPTDLVMVLTTAQWMTFTMKITTCAVYHIPILVTPKKRTVEHVIEIIENFRPTWTFFNPAFAKGLLAVVRREQIGSLETIILLGSLVTPDLVKEFKEKLHPDTHLCNGYGTTETQGFIAVTDRDAEASSNGWVYNILHYKLIDDNGAEVGPGSNGELYVKGECVVKGYYKNKEAYSESFVDGWYKTGDWFHLDENERLHFLERRKFNFKFRGCHVSPEEVEGVIGKLPGVHESVVVATEIGPAAAVVLLPEYDLTREDINKAVDSTLSDHKRLHGGIAFVDSLPHTHSGKLKRMECKKLIEELLKDGKCF
Summary
Uniprot
A0A2H1VL18
A0A0L7L9L4
A0A437BGV7
A0A194PRQ9
A0A0N0PC49
A0A0L7L8A5
+ More
A0A212FBS4 A0A1B3TP04 A0A1B3TNY4 A0A1B3TP14 A0A1B3TNZ3 A0A3S2N8I8 A0A1B3TNX4 A0A2W1BJT9 A0A2A4JFU1 A0A2A4JE16 A0A2H1VAR1 A0A2H1VV91 A0A194Q640 A0A194PZY0 A0A2W1BLY3 A0A2W1BGF2 A0A2A4J4G8 A0A194R1K6 A0A2A4J5M1 A0A1B3TP12 A0A194Q1K5 A0A212FN15 A0A1B3TNZ0 A0A1B3TNX9 A0A194PZW6 A0A2A4JKH2 A0A1B3TP22 A0A1B3TNZ4 A0A1B3TP07 A0A2W1BJK0 A0A194R232 A0A194Q0P0 A0A2H1V0G3 A0A2A4JCF3 A0A1B3TNZ2 A0A2A4JDK9 A0A1B3TNX2 A0A1B3TP24 A0A2H1VFT6 A0A1B3TP11 A0A1B3TNX3 A0A2H1W204 A0A2H1V5T4 A0A2H1V5V8 A0A1B3TNY0 A0A1B3TP25 A0A1B3TP01 A0A2H1VH10 A0A1B3TP10 A0A1B3TNW9 A0A2H1WMC6 A0A182GBF4 A0A2H1VGY5 A0A1B3TNZ6 A0A1B3TNZ5 E0W0Q3 A0A2H1V297 A0A2A4JUE1 A0A182L013 A0A1S4FRI6 A0A1S4GZC1 Q16RT8 A0A1B3TNX0 A0A2Y9D141 A0A2Y9D3J6 B0X7N6 A0A3S2NR21 Q174Q7 A0A182L014 A0A1I8M9U3 A0A1S4HCX9 A0A1B3TP16 A0A1S4FW35 A0A182P4M6 B0XJH9 A0A336L8I2 A0A1B3TP03 Q16M43 A0A336L5T1 A0A3F2YSK3 A0A336M4T4 A0A336MHV9 A0A1B3TNY1 Q5TS94 A0A336L9V0 U5EUC1 A0A1Q3F7P1 A0A182IYX5 A0A3S2PGQ8
A0A212FBS4 A0A1B3TP04 A0A1B3TNY4 A0A1B3TP14 A0A1B3TNZ3 A0A3S2N8I8 A0A1B3TNX4 A0A2W1BJT9 A0A2A4JFU1 A0A2A4JE16 A0A2H1VAR1 A0A2H1VV91 A0A194Q640 A0A194PZY0 A0A2W1BLY3 A0A2W1BGF2 A0A2A4J4G8 A0A194R1K6 A0A2A4J5M1 A0A1B3TP12 A0A194Q1K5 A0A212FN15 A0A1B3TNZ0 A0A1B3TNX9 A0A194PZW6 A0A2A4JKH2 A0A1B3TP22 A0A1B3TNZ4 A0A1B3TP07 A0A2W1BJK0 A0A194R232 A0A194Q0P0 A0A2H1V0G3 A0A2A4JCF3 A0A1B3TNZ2 A0A2A4JDK9 A0A1B3TNX2 A0A1B3TP24 A0A2H1VFT6 A0A1B3TP11 A0A1B3TNX3 A0A2H1W204 A0A2H1V5T4 A0A2H1V5V8 A0A1B3TNY0 A0A1B3TP25 A0A1B3TP01 A0A2H1VH10 A0A1B3TP10 A0A1B3TNW9 A0A2H1WMC6 A0A182GBF4 A0A2H1VGY5 A0A1B3TNZ6 A0A1B3TNZ5 E0W0Q3 A0A2H1V297 A0A2A4JUE1 A0A182L013 A0A1S4FRI6 A0A1S4GZC1 Q16RT8 A0A1B3TNX0 A0A2Y9D141 A0A2Y9D3J6 B0X7N6 A0A3S2NR21 Q174Q7 A0A182L014 A0A1I8M9U3 A0A1S4HCX9 A0A1B3TP16 A0A1S4FW35 A0A182P4M6 B0XJH9 A0A336L8I2 A0A1B3TP03 Q16M43 A0A336L5T1 A0A3F2YSK3 A0A336M4T4 A0A336MHV9 A0A1B3TNY1 Q5TS94 A0A336L9V0 U5EUC1 A0A1Q3F7P1 A0A182IYX5 A0A3S2PGQ8
Pubmed
EMBL
ODYU01003146
SOQ41530.1
JTDY01002178
KOB71941.1
RSAL01000060
RVE49662.1
+ More
KQ459595 KPI95997.1 KQ460652 KPJ12977.1 JTDY01002284 KOB71743.1 AGBW02009293 OWR51173.1 KU925757 AOG75382.1 KU925729 AOG75354.1 KU925772 AOG75397.1 KU925742 AOG75367.1 RSAL01000205 RVE44356.1 KU925719 AOG75344.1 KZ150128 PZC73096.1 NWSH01001632 PCG70636.1 NWSH01001926 PCG69652.1 ODYU01001291 SOQ37304.1 ODYU01004651 SOQ44755.1 KQ459582 KPI98875.1 KPI98876.1 KZ150081 PZC73876.1 PZC73878.1 NWSH01003054 PCG67037.1 KQ460878 KPJ11572.1 PCG67036.1 KU925758 AOG75383.1 KPI98874.1 AGBW02007648 OWR55128.1 KU925745 AOG75370.1 KU925730 AOG75355.1 KPI98872.1 NWSH01001271 PCG71900.1 KU925773 AOG75398.1 KU925743 AOG75368.1 KU925761 AOG75386.1 PZC73875.1 KPJ11574.1 KPI98873.1 ODYU01000111 SOQ34327.1 NWSH01001893 PCG69777.1 KU925733 AOG75358.1 PCG69778.1 KU925720 AOG75345.1 KU925775 AOG75400.1 ODYU01002332 SOQ39697.1 KU925759 AOG75384.1 KU925722 AOG75347.1 ODYU01005841 SOQ47125.1 ODYU01000839 SOQ36210.1 ODYU01000840 SOQ36211.1 KU925732 AOG75357.1 KU925774 AOG75399.1 KU925744 AOG75369.1 ODYU01002515 SOQ40108.1 KU925762 AOG75387.1 KU925721 AOG75346.1 ODYU01009613 SOQ54167.1 JXUM01008996 KQ560274 KXJ83386.1 SOQ40110.1 KU925747 AOG75372.1 KU925746 AOG75371.1 DS235862 EEB19209.1 ODYU01000351 SOQ34963.1 NWSH01000564 PCG75621.1 AAAB01008944 CH477697 EAT37103.1 KU925717 AOG75342.1 APCN01000732 DS232456 EDS42048.1 RSAL01000342 RVE42388.1 CH477407 EAT41541.1 KU925770 AOG75395.1 DS233521 EDS30382.1 UFQS01001959 UFQT01001959 SSX12785.1 SSX32227.1 KU925755 AOG75380.1 CH477881 EAT35397.1 SSX12784.1 SSX32226.1 APCN01000731 UFQS01000158 UFQT01000158 SSX00648.1 SSX21028.1 UFQS01001030 UFQT01001030 SSX08633.1 SSX28549.1 KU925740 AOG75365.1 EAL40219.3 UFQS01002704 UFQT01002704 SSX14568.1 SSX33966.1 GANO01001547 JAB58324.1 GFDL01011454 JAV23591.1 RSAL01000040 RVE50945.1
KQ459595 KPI95997.1 KQ460652 KPJ12977.1 JTDY01002284 KOB71743.1 AGBW02009293 OWR51173.1 KU925757 AOG75382.1 KU925729 AOG75354.1 KU925772 AOG75397.1 KU925742 AOG75367.1 RSAL01000205 RVE44356.1 KU925719 AOG75344.1 KZ150128 PZC73096.1 NWSH01001632 PCG70636.1 NWSH01001926 PCG69652.1 ODYU01001291 SOQ37304.1 ODYU01004651 SOQ44755.1 KQ459582 KPI98875.1 KPI98876.1 KZ150081 PZC73876.1 PZC73878.1 NWSH01003054 PCG67037.1 KQ460878 KPJ11572.1 PCG67036.1 KU925758 AOG75383.1 KPI98874.1 AGBW02007648 OWR55128.1 KU925745 AOG75370.1 KU925730 AOG75355.1 KPI98872.1 NWSH01001271 PCG71900.1 KU925773 AOG75398.1 KU925743 AOG75368.1 KU925761 AOG75386.1 PZC73875.1 KPJ11574.1 KPI98873.1 ODYU01000111 SOQ34327.1 NWSH01001893 PCG69777.1 KU925733 AOG75358.1 PCG69778.1 KU925720 AOG75345.1 KU925775 AOG75400.1 ODYU01002332 SOQ39697.1 KU925759 AOG75384.1 KU925722 AOG75347.1 ODYU01005841 SOQ47125.1 ODYU01000839 SOQ36210.1 ODYU01000840 SOQ36211.1 KU925732 AOG75357.1 KU925774 AOG75399.1 KU925744 AOG75369.1 ODYU01002515 SOQ40108.1 KU925762 AOG75387.1 KU925721 AOG75346.1 ODYU01009613 SOQ54167.1 JXUM01008996 KQ560274 KXJ83386.1 SOQ40110.1 KU925747 AOG75372.1 KU925746 AOG75371.1 DS235862 EEB19209.1 ODYU01000351 SOQ34963.1 NWSH01000564 PCG75621.1 AAAB01008944 CH477697 EAT37103.1 KU925717 AOG75342.1 APCN01000732 DS232456 EDS42048.1 RSAL01000342 RVE42388.1 CH477407 EAT41541.1 KU925770 AOG75395.1 DS233521 EDS30382.1 UFQS01001959 UFQT01001959 SSX12785.1 SSX32227.1 KU925755 AOG75380.1 CH477881 EAT35397.1 SSX12784.1 SSX32226.1 APCN01000731 UFQS01000158 UFQT01000158 SSX00648.1 SSX21028.1 UFQS01001030 UFQT01001030 SSX08633.1 SSX28549.1 KU925740 AOG75365.1 EAL40219.3 UFQS01002704 UFQT01002704 SSX14568.1 SSX33966.1 GANO01001547 JAB58324.1 GFDL01011454 JAV23591.1 RSAL01000040 RVE50945.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VL18
A0A0L7L9L4
A0A437BGV7
A0A194PRQ9
A0A0N0PC49
A0A0L7L8A5
+ More
A0A212FBS4 A0A1B3TP04 A0A1B3TNY4 A0A1B3TP14 A0A1B3TNZ3 A0A3S2N8I8 A0A1B3TNX4 A0A2W1BJT9 A0A2A4JFU1 A0A2A4JE16 A0A2H1VAR1 A0A2H1VV91 A0A194Q640 A0A194PZY0 A0A2W1BLY3 A0A2W1BGF2 A0A2A4J4G8 A0A194R1K6 A0A2A4J5M1 A0A1B3TP12 A0A194Q1K5 A0A212FN15 A0A1B3TNZ0 A0A1B3TNX9 A0A194PZW6 A0A2A4JKH2 A0A1B3TP22 A0A1B3TNZ4 A0A1B3TP07 A0A2W1BJK0 A0A194R232 A0A194Q0P0 A0A2H1V0G3 A0A2A4JCF3 A0A1B3TNZ2 A0A2A4JDK9 A0A1B3TNX2 A0A1B3TP24 A0A2H1VFT6 A0A1B3TP11 A0A1B3TNX3 A0A2H1W204 A0A2H1V5T4 A0A2H1V5V8 A0A1B3TNY0 A0A1B3TP25 A0A1B3TP01 A0A2H1VH10 A0A1B3TP10 A0A1B3TNW9 A0A2H1WMC6 A0A182GBF4 A0A2H1VGY5 A0A1B3TNZ6 A0A1B3TNZ5 E0W0Q3 A0A2H1V297 A0A2A4JUE1 A0A182L013 A0A1S4FRI6 A0A1S4GZC1 Q16RT8 A0A1B3TNX0 A0A2Y9D141 A0A2Y9D3J6 B0X7N6 A0A3S2NR21 Q174Q7 A0A182L014 A0A1I8M9U3 A0A1S4HCX9 A0A1B3TP16 A0A1S4FW35 A0A182P4M6 B0XJH9 A0A336L8I2 A0A1B3TP03 Q16M43 A0A336L5T1 A0A3F2YSK3 A0A336M4T4 A0A336MHV9 A0A1B3TNY1 Q5TS94 A0A336L9V0 U5EUC1 A0A1Q3F7P1 A0A182IYX5 A0A3S2PGQ8
A0A212FBS4 A0A1B3TP04 A0A1B3TNY4 A0A1B3TP14 A0A1B3TNZ3 A0A3S2N8I8 A0A1B3TNX4 A0A2W1BJT9 A0A2A4JFU1 A0A2A4JE16 A0A2H1VAR1 A0A2H1VV91 A0A194Q640 A0A194PZY0 A0A2W1BLY3 A0A2W1BGF2 A0A2A4J4G8 A0A194R1K6 A0A2A4J5M1 A0A1B3TP12 A0A194Q1K5 A0A212FN15 A0A1B3TNZ0 A0A1B3TNX9 A0A194PZW6 A0A2A4JKH2 A0A1B3TP22 A0A1B3TNZ4 A0A1B3TP07 A0A2W1BJK0 A0A194R232 A0A194Q0P0 A0A2H1V0G3 A0A2A4JCF3 A0A1B3TNZ2 A0A2A4JDK9 A0A1B3TNX2 A0A1B3TP24 A0A2H1VFT6 A0A1B3TP11 A0A1B3TNX3 A0A2H1W204 A0A2H1V5T4 A0A2H1V5V8 A0A1B3TNY0 A0A1B3TP25 A0A1B3TP01 A0A2H1VH10 A0A1B3TP10 A0A1B3TNW9 A0A2H1WMC6 A0A182GBF4 A0A2H1VGY5 A0A1B3TNZ6 A0A1B3TNZ5 E0W0Q3 A0A2H1V297 A0A2A4JUE1 A0A182L013 A0A1S4FRI6 A0A1S4GZC1 Q16RT8 A0A1B3TNX0 A0A2Y9D141 A0A2Y9D3J6 B0X7N6 A0A3S2NR21 Q174Q7 A0A182L014 A0A1I8M9U3 A0A1S4HCX9 A0A1B3TP16 A0A1S4FW35 A0A182P4M6 B0XJH9 A0A336L8I2 A0A1B3TP03 Q16M43 A0A336L5T1 A0A3F2YSK3 A0A336M4T4 A0A336MHV9 A0A1B3TNY1 Q5TS94 A0A336L9V0 U5EUC1 A0A1Q3F7P1 A0A182IYX5 A0A3S2PGQ8
PDB
6AC3
E-value=8.45332e-35,
Score=370
Ontologies
GO
Topology
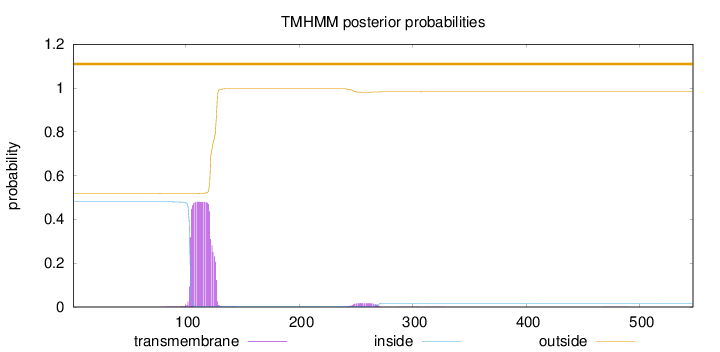
Length:
547
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.40985
Exp number, first 60 AAs:
0.00072
Total prob of N-in:
0.48071
outside
1 - 547
Population Genetic Test Statistics
Pi
283.040714
Theta
175.838028
Tajima's D
1.838103
CLR
0.226169
CSRT
0.853157342132893
Interpretation
Uncertain
