Gene
KWMTBOMO09863
Pre Gene Modal
BGIBMGA013156
Annotation
PREDICTED:_TWiK_family_of_potassium_channels_protein_7-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.197
Sequence
CDS
ATGCAAGTTTTATGGAAGAACCGAAATGCTGTTTTACAAGAAGAGGAGAACTGGACCAAATTAGCTGCTAAAGAGTTAATGGATTTCCAAAAGGTTCTTATAAAGAGCATGCGCGGCGGCGTACTGTTGGACAATAAGCTAACCTTTGATCACGAATCCGACTACAGATGGACATTTTCAAATAGTTTCCTTTATGCCTTGACTTTGATCACGACGATTGGCCATGGAAACGTCACTCCTAAATCATCAGTGGGGAAAATGGTTGCTGTGGTCTATGCTTGCATCGGCATTCCCATAATTATGCTGTATCTCTCCACTATCGGCGAAGCACTGGCCCGAAACTTCCGAGTGCTTTATTCGAAGATCTGCCCATCGAGCCAGAACAGCGACAGCTTCCACACCAAAGCAGACTGCTTGAACCGACACTCTGTTCAAGGCGAGTGCAGTAAAGATAAAAGGGAGAAGTACAGTGGCGAACAACAAAAGCGGACCCCTTTTCAAGCTGCGTTAAATTTAGATAATTTTAGTGGTGGCTATCATTGGACGTGTCGGGACCACACTCGTGTGCCAATTATTCTTAGTTTCTTTCTAATCGTACTTTATATAGGATTAGGCTCCTTTGTATTCCACACGACAGAGAAATGGGACTTCATGGACGGATGTTTCTTTTCATTCGCAAGTTTGGCAACCATAGGTTTTGGTGACTTGAAACCAGGGTTGTATGCCAGCACCGTTTCTCCCAAGGCTGAAGATATAGCTGTAGGCGTGTGCTGCATTTACATTTTAGTGGGTATCGTAATAATAGCTATGTGTTTTAATCTCATACAAGAAGACATGAGTACGATGTTAAAGAAATTTACGACATTATGTACGGGAACTGCGAAATCTCGAGCCGTGCACAGCGTGGAGAAGATATGCGATGATAAAATACCTATGTCAATTGTTTCCTGA
Protein
MQVLWKNRNAVLQEEENWTKLAAKELMDFQKVLIKSMRGGVLLDNKLTFDHESDYRWTFSNSFLYALTLITTIGHGNVTPKSSVGKMVAVVYACIGIPIIMLYLSTIGEALARNFRVLYSKICPSSQNSDSFHTKADCLNRHSVQGECSKDKREKYSGEQQKRTPFQAALNLDNFSGGYHWTCRDHTRVPIILSFFLIVLYIGLGSFVFHTTEKWDFMDGCFFSFASLATIGFGDLKPGLYASTVSPKAEDIAVGVCCIYILVGIVIIAMCFNLIQEDMSTMLKKFTTLCTGTAKSRAVHSVEKICDDKIPMSIVS
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
A0A3S2N978
A0A2H1W8J8
A0A194PXX7
A0A0L7L1Z8
A0A212FM49
A0A0N1I6X3
+ More
A0A2S2NIS9 A0A1B6BXP2 A0A2H8TJ61 A0A1B6DHS3 A0A0A9Z638 A0A2L2YQC8 A0A182R2U0 A0A182K5F1 A0A2R7WRI0 A0A453YR86 A0A182XB81 A0A1S4GMN2 A0A182HWZ6 A0A182WAT8 A0A182PNG0 A0A182NN86 A0A182FLS6 A0A182GEM1 D2A3Q6 V3ZUS1 A0A0M4E3U7 A0A1J1HUR8 E2C7D3 A0A2J7Q9D4 T1IJW1 A0A1B6I5R3 A0A067QMW6 A0A1J1HUR2 D2A6I4 A0A1J1HWK4 A0A154PA07 V3ZRN1 A0A1I8PU96 A0A2S2NE42 A0A2H8TLH3 A0A3L8DE05 R7U6K0 A0A0L8GLX0 A0A182QHT4 A0A210QTX8 A0A1B6DH55 A0A1B6LPX4 T1EGS2 A0A0P4Y6Y5 A0A0M3QZE2 E9GA12 A0A182YI69 A0A182NN88 K1PKK9 B4L2G2 A0A182R2U3 A0A158NJU8 R7U1G0 A0A226E6Z7 T1EJ43 A0A044V410 T1G0W3 A0A1D1VS01 E3LS47 A0A1I7VTG2 A0A1D1VQB7 D6WYP0 A0A139WDM6 A0A074ZT08 A0A1S0U587 A0A2P6K3W2 E9H402 A0A2P6KZF1 R7UC91 G7YHV4 A0A3R7ERI4 A0A0B2UMR5 A0A154PFT0 A0A3M7QG93 A0A2R4KQY6 A0A2A6CIP7 A0A195FSD6 A0A183ALE8 A0A0L7RCB2 A0A1W4X1N0 A0A0N8BSJ7 A0A0P5DCC5 A0A0P5SAB2 A0A0P5BE98 A0A0P5Q7G1 E9H9W3 T1EDB5 A0A0P5J1L8 A0A139WC92
A0A2S2NIS9 A0A1B6BXP2 A0A2H8TJ61 A0A1B6DHS3 A0A0A9Z638 A0A2L2YQC8 A0A182R2U0 A0A182K5F1 A0A2R7WRI0 A0A453YR86 A0A182XB81 A0A1S4GMN2 A0A182HWZ6 A0A182WAT8 A0A182PNG0 A0A182NN86 A0A182FLS6 A0A182GEM1 D2A3Q6 V3ZUS1 A0A0M4E3U7 A0A1J1HUR8 E2C7D3 A0A2J7Q9D4 T1IJW1 A0A1B6I5R3 A0A067QMW6 A0A1J1HUR2 D2A6I4 A0A1J1HWK4 A0A154PA07 V3ZRN1 A0A1I8PU96 A0A2S2NE42 A0A2H8TLH3 A0A3L8DE05 R7U6K0 A0A0L8GLX0 A0A182QHT4 A0A210QTX8 A0A1B6DH55 A0A1B6LPX4 T1EGS2 A0A0P4Y6Y5 A0A0M3QZE2 E9GA12 A0A182YI69 A0A182NN88 K1PKK9 B4L2G2 A0A182R2U3 A0A158NJU8 R7U1G0 A0A226E6Z7 T1EJ43 A0A044V410 T1G0W3 A0A1D1VS01 E3LS47 A0A1I7VTG2 A0A1D1VQB7 D6WYP0 A0A139WDM6 A0A074ZT08 A0A1S0U587 A0A2P6K3W2 E9H402 A0A2P6KZF1 R7UC91 G7YHV4 A0A3R7ERI4 A0A0B2UMR5 A0A154PFT0 A0A3M7QG93 A0A2R4KQY6 A0A2A6CIP7 A0A195FSD6 A0A183ALE8 A0A0L7RCB2 A0A1W4X1N0 A0A0N8BSJ7 A0A0P5DCC5 A0A0P5SAB2 A0A0P5BE98 A0A0P5Q7G1 E9H9W3 T1EDB5 A0A0P5J1L8 A0A139WC92
Pubmed
EMBL
RSAL01000497
RVE41529.1
ODYU01006903
SOQ49172.1
KQ459595
KPI96000.1
+ More
JTDY01003458 KOB69528.1 AGBW02007655 OWR54811.1 KQ460652 KPJ12973.1 GGMR01004465 MBY17084.1 GEDC01031518 GEDC01025482 GEDC01019003 JAS05780.1 JAS11816.1 JAS18295.1 GFXV01002341 MBW14146.1 GEDC01026712 GEDC01014941 GEDC01012080 JAS10586.1 JAS22357.1 JAS25218.1 GBHO01025226 GBHO01025221 GBHO01004258 GBHO01004257 GBHO01004255 JAG18378.1 JAG18383.1 JAG39346.1 JAG39347.1 JAG39349.1 IAAA01031791 IAAA01031792 LAA09565.1 KK855358 PTY22164.1 AAAB01008968 APCN01006853 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1 KQ971348 EFA04896.1 KB203440 ESO84681.1 CP012524 ALC40566.1 CVRI01000021 CRK91749.1 GL453369 EFN76120.1 NEVH01016370 PNF25195.1 JH430359 GECU01025447 JAS82259.1 KK853235 KDR09567.1 CRK91739.1 EFA04915.1 CRK91740.1 KQ434839 KZC08020.1 KB202954 ESO86997.1 GGMR01002790 MBY15409.1 GFXV01003180 MBW14985.1 QOIP01000009 RLU18665.1 AMQN01001996 KB307554 ELT98770.1 KQ421376 KOF77590.1 AXCN02001900 NEDP02001925 OWF52167.1 GEDC01012257 JAS25041.1 GEBQ01014187 JAT25790.1 AMQM01007336 KB097599 ESN93612.1 GDIP01233930 JAI89471.1 CP012528 ALC49219.1 GL732536 EFX83663.1 JH817993 EKC24562.1 CH933810 EDW06838.2 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 AMQN01009906 KB306576 ELT99712.1 LNIX01000006 OXA52737.1 AMQM01003437 KB096134 ESO08197.1 CMVM020000249 AMQM01002765 KB095858 ESO10860.1 BDGG01000010 GAV03751.1 DS268414 EFP09568.1 GAV03750.1 KQ971357 EFA08415.2 KYB26059.1 KL596664 KER30236.1 JH712223 EFO25445.2 MWRG01031807 PRD21016.1 GL732589 EFX73616.1 MWRG01003347 PRD31706.1 AMQN01009528 KB305619 ELU00882.1 DF143310 GAA52537.1 NIRI01002155 RJW63468.1 JPKZ01003266 KHN72276.1 KQ434893 KZC10672.1 REGN01006254 RNA10242.1 MF069081 AVV64028.1 ABKE03000028 PDM77947.1 KQ981305 KYN42814.1 UZAN01045073 VDP82008.1 KQ414616 KOC68464.1 GDIQ01149083 JAL02643.1 GDIP01162143 JAJ61259.1 GDIQ01098888 LRGB01001361 JAL52838.1 KZS12615.1 GDIP01186434 JAJ36968.1 GDIQ01118089 JAL33637.1 GL732609 EFX71471.1 AMQM01006994 KB097552 ESN94959.1 GDIQ01204398 JAK47327.1 KQ971372 KYB25451.1
JTDY01003458 KOB69528.1 AGBW02007655 OWR54811.1 KQ460652 KPJ12973.1 GGMR01004465 MBY17084.1 GEDC01031518 GEDC01025482 GEDC01019003 JAS05780.1 JAS11816.1 JAS18295.1 GFXV01002341 MBW14146.1 GEDC01026712 GEDC01014941 GEDC01012080 JAS10586.1 JAS22357.1 JAS25218.1 GBHO01025226 GBHO01025221 GBHO01004258 GBHO01004257 GBHO01004255 JAG18378.1 JAG18383.1 JAG39346.1 JAG39347.1 JAG39349.1 IAAA01031791 IAAA01031792 LAA09565.1 KK855358 PTY22164.1 AAAB01008968 APCN01006853 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1 KQ971348 EFA04896.1 KB203440 ESO84681.1 CP012524 ALC40566.1 CVRI01000021 CRK91749.1 GL453369 EFN76120.1 NEVH01016370 PNF25195.1 JH430359 GECU01025447 JAS82259.1 KK853235 KDR09567.1 CRK91739.1 EFA04915.1 CRK91740.1 KQ434839 KZC08020.1 KB202954 ESO86997.1 GGMR01002790 MBY15409.1 GFXV01003180 MBW14985.1 QOIP01000009 RLU18665.1 AMQN01001996 KB307554 ELT98770.1 KQ421376 KOF77590.1 AXCN02001900 NEDP02001925 OWF52167.1 GEDC01012257 JAS25041.1 GEBQ01014187 JAT25790.1 AMQM01007336 KB097599 ESN93612.1 GDIP01233930 JAI89471.1 CP012528 ALC49219.1 GL732536 EFX83663.1 JH817993 EKC24562.1 CH933810 EDW06838.2 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 AMQN01009906 KB306576 ELT99712.1 LNIX01000006 OXA52737.1 AMQM01003437 KB096134 ESO08197.1 CMVM020000249 AMQM01002765 KB095858 ESO10860.1 BDGG01000010 GAV03751.1 DS268414 EFP09568.1 GAV03750.1 KQ971357 EFA08415.2 KYB26059.1 KL596664 KER30236.1 JH712223 EFO25445.2 MWRG01031807 PRD21016.1 GL732589 EFX73616.1 MWRG01003347 PRD31706.1 AMQN01009528 KB305619 ELU00882.1 DF143310 GAA52537.1 NIRI01002155 RJW63468.1 JPKZ01003266 KHN72276.1 KQ434893 KZC10672.1 REGN01006254 RNA10242.1 MF069081 AVV64028.1 ABKE03000028 PDM77947.1 KQ981305 KYN42814.1 UZAN01045073 VDP82008.1 KQ414616 KOC68464.1 GDIQ01149083 JAL02643.1 GDIP01162143 JAJ61259.1 GDIQ01098888 LRGB01001361 JAL52838.1 KZS12615.1 GDIP01186434 JAJ36968.1 GDIQ01118089 JAL33637.1 GL732609 EFX71471.1 AMQM01006994 KB097552 ESN94959.1 GDIQ01204398 JAK47327.1 KQ971372 KYB25451.1
Proteomes
UP000283053
UP000053268
UP000037510
UP000007151
UP000053240
UP000075900
+ More
UP000075881 UP000076407 UP000075840 UP000075920 UP000075885 UP000075884 UP000069272 UP000069940 UP000249989 UP000007266 UP000030746 UP000092553 UP000183832 UP000008237 UP000235965 UP000027135 UP000076502 UP000095300 UP000279307 UP000014760 UP000053454 UP000075886 UP000242188 UP000015101 UP000000305 UP000076408 UP000005408 UP000009192 UP000005205 UP000198287 UP000024404 UP000186922 UP000008281 UP000095285 UP000031036 UP000276133 UP000232936 UP000078541 UP000050740 UP000053825 UP000192223 UP000076858
UP000075881 UP000076407 UP000075840 UP000075920 UP000075885 UP000075884 UP000069272 UP000069940 UP000249989 UP000007266 UP000030746 UP000092553 UP000183832 UP000008237 UP000235965 UP000027135 UP000076502 UP000095300 UP000279307 UP000014760 UP000053454 UP000075886 UP000242188 UP000015101 UP000000305 UP000076408 UP000005408 UP000009192 UP000005205 UP000198287 UP000024404 UP000186922 UP000008281 UP000095285 UP000031036 UP000276133 UP000232936 UP000078541 UP000050740 UP000053825 UP000192223 UP000076858
Pfam
PF07885 Ion_trans_2
ProteinModelPortal
A0A3S2N978
A0A2H1W8J8
A0A194PXX7
A0A0L7L1Z8
A0A212FM49
A0A0N1I6X3
+ More
A0A2S2NIS9 A0A1B6BXP2 A0A2H8TJ61 A0A1B6DHS3 A0A0A9Z638 A0A2L2YQC8 A0A182R2U0 A0A182K5F1 A0A2R7WRI0 A0A453YR86 A0A182XB81 A0A1S4GMN2 A0A182HWZ6 A0A182WAT8 A0A182PNG0 A0A182NN86 A0A182FLS6 A0A182GEM1 D2A3Q6 V3ZUS1 A0A0M4E3U7 A0A1J1HUR8 E2C7D3 A0A2J7Q9D4 T1IJW1 A0A1B6I5R3 A0A067QMW6 A0A1J1HUR2 D2A6I4 A0A1J1HWK4 A0A154PA07 V3ZRN1 A0A1I8PU96 A0A2S2NE42 A0A2H8TLH3 A0A3L8DE05 R7U6K0 A0A0L8GLX0 A0A182QHT4 A0A210QTX8 A0A1B6DH55 A0A1B6LPX4 T1EGS2 A0A0P4Y6Y5 A0A0M3QZE2 E9GA12 A0A182YI69 A0A182NN88 K1PKK9 B4L2G2 A0A182R2U3 A0A158NJU8 R7U1G0 A0A226E6Z7 T1EJ43 A0A044V410 T1G0W3 A0A1D1VS01 E3LS47 A0A1I7VTG2 A0A1D1VQB7 D6WYP0 A0A139WDM6 A0A074ZT08 A0A1S0U587 A0A2P6K3W2 E9H402 A0A2P6KZF1 R7UC91 G7YHV4 A0A3R7ERI4 A0A0B2UMR5 A0A154PFT0 A0A3M7QG93 A0A2R4KQY6 A0A2A6CIP7 A0A195FSD6 A0A183ALE8 A0A0L7RCB2 A0A1W4X1N0 A0A0N8BSJ7 A0A0P5DCC5 A0A0P5SAB2 A0A0P5BE98 A0A0P5Q7G1 E9H9W3 T1EDB5 A0A0P5J1L8 A0A139WC92
A0A2S2NIS9 A0A1B6BXP2 A0A2H8TJ61 A0A1B6DHS3 A0A0A9Z638 A0A2L2YQC8 A0A182R2U0 A0A182K5F1 A0A2R7WRI0 A0A453YR86 A0A182XB81 A0A1S4GMN2 A0A182HWZ6 A0A182WAT8 A0A182PNG0 A0A182NN86 A0A182FLS6 A0A182GEM1 D2A3Q6 V3ZUS1 A0A0M4E3U7 A0A1J1HUR8 E2C7D3 A0A2J7Q9D4 T1IJW1 A0A1B6I5R3 A0A067QMW6 A0A1J1HUR2 D2A6I4 A0A1J1HWK4 A0A154PA07 V3ZRN1 A0A1I8PU96 A0A2S2NE42 A0A2H8TLH3 A0A3L8DE05 R7U6K0 A0A0L8GLX0 A0A182QHT4 A0A210QTX8 A0A1B6DH55 A0A1B6LPX4 T1EGS2 A0A0P4Y6Y5 A0A0M3QZE2 E9GA12 A0A182YI69 A0A182NN88 K1PKK9 B4L2G2 A0A182R2U3 A0A158NJU8 R7U1G0 A0A226E6Z7 T1EJ43 A0A044V410 T1G0W3 A0A1D1VS01 E3LS47 A0A1I7VTG2 A0A1D1VQB7 D6WYP0 A0A139WDM6 A0A074ZT08 A0A1S0U587 A0A2P6K3W2 E9H402 A0A2P6KZF1 R7UC91 G7YHV4 A0A3R7ERI4 A0A0B2UMR5 A0A154PFT0 A0A3M7QG93 A0A2R4KQY6 A0A2A6CIP7 A0A195FSD6 A0A183ALE8 A0A0L7RCB2 A0A1W4X1N0 A0A0N8BSJ7 A0A0P5DCC5 A0A0P5SAB2 A0A0P5BE98 A0A0P5Q7G1 E9H9W3 T1EDB5 A0A0P5J1L8 A0A139WC92
PDB
6CQ9
E-value=1.34895e-08,
Score=141
Ontologies
GO
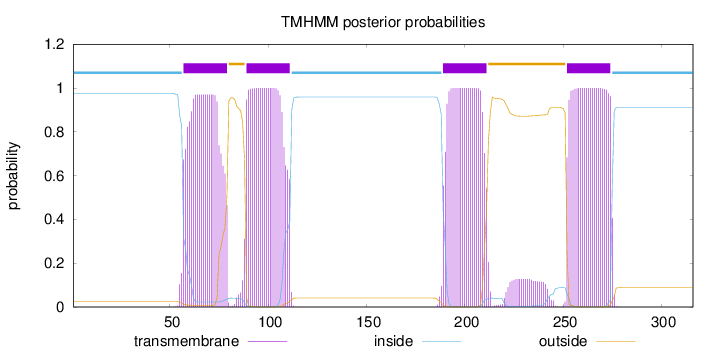
Topology
Length:
316
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.5110200000001
Exp number, first 60 AAs:
3.33267
Total prob of N-in:
0.97454
inside
1 - 56
TMhelix
57 - 79
outside
80 - 88
TMhelix
89 - 111
inside
112 - 188
TMhelix
189 - 211
outside
212 - 251
TMhelix
252 - 274
inside
275 - 316
Population Genetic Test Statistics
Pi
32.216882
Theta
164.257812
Tajima's D
-1.330574
CLR
1455.93681
CSRT
0.0843957802109895
Interpretation
Possibly Positive selection
