Gene
KWMTBOMO09860
Pre Gene Modal
BGIBMGA013155
Annotation
PREDICTED:_fatty_acid_synthase-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.088
Sequence
CDS
ATGATGTACCCTATAGGCGGAATATTAATGCTGGTATGGGATACACTTTCTATGTCAATGTCTTTGCCTAAGAGGAAAATATCGGTTAGATTTCTCGATTTATTTATCGATGTTCAACCAGCACTTTACGAAGAAAAAATATTAAAATTGCGCGTCGCGATCAATAAGGGCGATGGACAATTTGAGGTTACAAATGATGGATCCAAAGTTGCAAGTGGAATTGTTTTACTGGTTAGCAAAAATATATTCGGAAAAGAAACTCATACAACTGACACTCCAATGACCTTGAACACGAATGACTTCTATCAACTACTACAAGAAAAAGGATACTCTTACAGCGAAGATTTCCGTCTGATAAAGAGTGTTAACTTGTCTTTATCAGAAGCCATGATAGAATGGCGCGATAACTGGGTGACGTTGATTGAGAATATGCTGCAGCTTGTTATCCTGAATCAGAACTATGATGGTGTTTTACAACCAACTCAGATTAATAGAATTGTTATCGATACCATTGAATCTTCAAATCAAACTTTTACAAAGCATGGAAAATGCGTATATATTAAGGATATTCAATTCCGTGAAACTTCTCGAACTCCATATCCCATTAGTCTGTTAGTTCCGTCGAGTGAAGAAAAAGGAAAATTGATTAACTTCAAAAGTGTGACTAGCTCATACACAAATGCGATGCTCCAAAGCGCGGAAACTGGGAACGTTAATTCCTTGCACTGGGCTGAAGTTTCTGAACCAAGTGGATCTGGAGTACTTGTTAAGGTACATTACGCAAGTCTTTCAAATTACGACGGTAAGAAGGCCTCAGGAACAGTACCGCATCGAGCCAGCGAGAACTATTATGGAATAGACTTCAGTGGGATAGCCGAAAGCGGAAGTCGTGTGATGGGTATAGTTCATCGTGGTGCTTGTCGAAATTATGTTCGAGCTCACCCCGATCTTTTGTGGCCAGTCCCTGATCACTGGAGCCTCGAAGACGCTGCCACAGTTCCTTTAGCTTACGCGTATGCTTTTTATTGTCTAGCAATCAAAGCTAAGCTGTGCGCCGGTATGAAAATACTTATACACGAAGGAGCTGGAGCACTCGGCCAGGCTGTCATTGCCATCGCTCTGGCACACAAATGCCAAGTATTCACTACAGTAGGGGATTTAAGAACGAAAAGGTTCTTGAAGAAACTATTCCCAGAATTGAAAGAGGAACATATTGGAAATTCTAGAGACATCACTTTCGGGGACATGGTTCTAACAGCGACAAATGGGAAAGGTTGCGATATTATAATAAGCAATGTCGAAACATCACTCAAAAACGTTTCACTTCAATGCGGTAATTATCAGAGCGTTATTTTTGATACGGAACAAATTGAGAAACGCGAGAATTTCGAATTGCGTATGAACAGTCTGATTAATGACAGATCCTATGTAACTACTAAGTTCGAGTCAATTTTTGAATTTTGGAATTCTGAAGAAATCAAAAATTTACAAATACTTATGAATGAAGGCATCTCCAGTGGCTACGTTCGTCCATTAGCTAGAGTGTCATACTCTTCAAAGGAAGCTCCTCGTGCTTTTCGACTCTTATGCAAGAATTGGCATCGAGGACATATATTATTGAGACTCACAGAACAAATGCTGCCAGCTCAACCAAGGATCAGCTGTAGATCCGAATGCAGTCAGTTAATCATCTGCAGCGAAGAAGATTTCGGTATTCAGTTAGCGAACAGACTTGTCAGTAGAGGTGCTACAAAAATACACATTTGTAGTCCGAAAGTCTCATGGTATCTCCAACATAAAACAGACAATTTAAAATCTATTGGTGTCAATGTGGTCGTATCTACGAACATAATAAAGGATTCTTACGATACTACATTTCTTTTAAAAAAAACTATAGCTATGGGACCATTAGAGGGCATTTATGTGGCAAACTTTAGAAATGACGACGAAACATTAAACATCCTGCAAGACATTGGTAATATGTCCAAAGATTTATTTCCATCATTAAGGTACTTTGCAGTGATTACCAACAATGTAAAAGTTCTAGAGAAAACGTGTATTTCTGGAGTAAACGATAATTCAATTATTTCTCTAATCGAAACTCGAAGGCCGCAAGTAAATGAGAATGAAAAAGACGCCATCTTATTAGACAAGGGAGCTTGGAACTTATCCACATTGAACGCTTTAGAGACCGGACTTCGCTGCAAGGTCCCATTGTTGGTCGCCCAAGGAAACACTTACACATCACCGCTGTATCAACAACTTGCAGCTTCTGCTGATATTCCATCTTTAGAAGATGCAAACGACGTCACTACTCTACGAGAATTGTCTGTTGGAAGCGAAAACATTAAAATAGTTAAAGAATTTCTACGTCAAAAATACCATAAGTGCTTTTTGGAACAGGAAATTTTGGAGTTAAACAAGCAGAAGCTTGAAGAGTTGGATTCTCTTCAAAACGGCTTCGAGAAGAAAGAAGGCTTAGCTGTTTACTTGTCTCACATAGCTAAAGATGAACTCAATTCCAATGCCCTCGTGTACTCTTTACCCACACTTGGCAATACGACAACGGAGAAACTGTCTGAAGCAATTGAGTCCGAGTACTTGTGCATAGTTCTTGGGATCGAGAATAATTTCGAAAGGTTTTATGGTATTTGCGAGAGACTTAAGCTGCCCGCGCTAGTGCTGCAGCCACGTCTGGATAGAACAAGCGATACTATACAGCAAACCGCCGAAAAATGCTCAAAGTTGATAACAGAGCAAATTGGTGTCAGAAAAAGTTTTTACTTGCTCGCATCCGATATCGGTATTTTAATTGCCATGGAGTTAGCTGCAATATTTGAGAGTAATGGTATTTTAGGCACAGTCTTCTGTCTCGGTAGTGCACCAGAAGATGTCAAGCCAATAATTGAACGTGAACTAGCCGCCTTCAAAACCGAAGACCAACTGCAGCTTGAGATTTTAAAGCACATTTTTAAGCTACTCACCGCTAATGATCCATCAATTTTAGAGAAAGCTTTGCCTCCTTTGTCCTCATGGAATGAAAAGATAGAGGGTTGCATTAAAATGATGACTGGTCATATAGCACAGACAAAGCAATATGTGAAAGCTTTGATACAATCTGTCTATGCGAAAATTCTTAGCATCAAGCAGTATGATTCTTCAACTCTGCCAATATTGAAGTCTCGAGTCATCATACTCCGCTCTCCGGATTGCTCCATTACTTCATCCCCATCATCATATTCTATTGAAGATTTATCAGCACCTCTTAGCGACGCTGCAACAGATACGCGATGTGCAGCCATTATCAACAAAAATCTGTCGAGTGAAATATTAGAAGAGTTTGATAAGAAAAACCTCTGCGACACATTTTTGATAAGCTTTGATGAGAAATATACAAAAGTTAATTCTAGAGTAATATGA
Protein
MMYPIGGILMLVWDTLSMSMSLPKRKISVRFLDLFIDVQPALYEEKILKLRVAINKGDGQFEVTNDGSKVASGIVLLVSKNIFGKETHTTDTPMTLNTNDFYQLLQEKGYSYSEDFRLIKSVNLSLSEAMIEWRDNWVTLIENMLQLVILNQNYDGVLQPTQINRIVIDTIESSNQTFTKHGKCVYIKDIQFRETSRTPYPISLLVPSSEEKGKLINFKSVTSSYTNAMLQSAETGNVNSLHWAEVSEPSGSGVLVKVHYASLSNYDGKKASGTVPHRASENYYGIDFSGIAESGSRVMGIVHRGACRNYVRAHPDLLWPVPDHWSLEDAATVPLAYAYAFYCLAIKAKLCAGMKILIHEGAGALGQAVIAIALAHKCQVFTTVGDLRTKRFLKKLFPELKEEHIGNSRDITFGDMVLTATNGKGCDIIISNVETSLKNVSLQCGNYQSVIFDTEQIEKRENFELRMNSLINDRSYVTTKFESIFEFWNSEEIKNLQILMNEGISSGYVRPLARVSYSSKEAPRAFRLLCKNWHRGHILLRLTEQMLPAQPRISCRSECSQLIICSEEDFGIQLANRLVSRGATKIHICSPKVSWYLQHKTDNLKSIGVNVVVSTNIIKDSYDTTFLLKKTIAMGPLEGIYVANFRNDDETLNILQDIGNMSKDLFPSLRYFAVITNNVKVLEKTCISGVNDNSIISLIETRRPQVNENEKDAILLDKGAWNLSTLNALETGLRCKVPLLVAQGNTYTSPLYQQLAASADIPSLEDANDVTTLRELSVGSENIKIVKEFLRQKYHKCFLEQEILELNKQKLEELDSLQNGFEKKEGLAVYLSHIAKDELNSNALVYSLPTLGNTTTEKLSEAIESEYLCIVLGIENNFERFYGICERLKLPALVLQPRLDRTSDTIQQTAEKCSKLITEQIGVRKSFYLLASDIGILIAMELAAIFESNGILGTVFCLGSAPEDVKPIIERELAAFKTEDQLQLEILKHIFKLLTANDPSILEKALPPLSSWNEKIEGCIKMMTGHIAQTKQYVKALIQSVYAKILSIKQYDSSTLPILKSRVIILRSPDCSITSSPSSYSIEDLSAPLSDAATDTRCAAIINKNLSSEILEEFDKKNLCDTFLISFDEKYTKVNSRVI
Summary
Uniprot
A0A2A4K3Y0
A0A437B0L0
A0A194PKD7
A0A0N1I4Z6
A0A2A4K3Y5
A0A437BUA6
+ More
A0A2A4K5B4 A0A437BUJ8 A0A0L7L2W8 A0A212EN78 O01677 A0A0L7L1N0 A0A3S2NJM7 A0A2H1V933 H9JGF5 A0A2H1WZR1 A0A2A4JVR3 A0A0L7KUR1 A0A2A4J169 A0A1B6CGM1 A0A232EXC2 H9JU35 A0A154PMH1 O01678 K7IZC8 A0A2A3E4Z2 A0A0M8ZRC2 E2B583 A0A088ALX8 E2BR60 A0A212FMU5 A0A0J7NXT7 A0A310SRI2 E2ADN8 D2A4M7 A0A1B6JP24 A0A0C9R1I7 A0A0L7R328 A0A026WNV9 A0A195BMY6 A0A194Q6Q7 A0A2H1WKG4 A0A158NZW8 A0A2J7PQY0 D2A4M4 A0A2H1VFB6 K7IZC9 A0A151X6P9 A0A1Y1MBP7 A0A2H1W5A9 A0A1J1IKA4 E2BGH5 E0W0Z3 F4WIF1 A0A2P8XAG4 A0A2C9PHR5 U4UFT3 A0A182M3X3 A0A182VQY2 A0A195EVF8 A0A151X557 V5GRN6 A0A195CP13 A0A182J9B8 A0A151I5V4 A0A344X1T4 T1PGU1 N6TYU8 A0A195CLZ5 E2C0H8
A0A2A4K5B4 A0A437BUJ8 A0A0L7L2W8 A0A212EN78 O01677 A0A0L7L1N0 A0A3S2NJM7 A0A2H1V933 H9JGF5 A0A2H1WZR1 A0A2A4JVR3 A0A0L7KUR1 A0A2A4J169 A0A1B6CGM1 A0A232EXC2 H9JU35 A0A154PMH1 O01678 K7IZC8 A0A2A3E4Z2 A0A0M8ZRC2 E2B583 A0A088ALX8 E2BR60 A0A212FMU5 A0A0J7NXT7 A0A310SRI2 E2ADN8 D2A4M7 A0A1B6JP24 A0A0C9R1I7 A0A0L7R328 A0A026WNV9 A0A195BMY6 A0A194Q6Q7 A0A2H1WKG4 A0A158NZW8 A0A2J7PQY0 D2A4M4 A0A2H1VFB6 K7IZC9 A0A151X6P9 A0A1Y1MBP7 A0A2H1W5A9 A0A1J1IKA4 E2BGH5 E0W0Z3 F4WIF1 A0A2P8XAG4 A0A2C9PHR5 U4UFT3 A0A182M3X3 A0A182VQY2 A0A195EVF8 A0A151X557 V5GRN6 A0A195CP13 A0A182J9B8 A0A151I5V4 A0A344X1T4 T1PGU1 N6TYU8 A0A195CLZ5 E2C0H8
Pubmed
EMBL
NWSH01000156
PCG78965.1
RSAL01000223
RVE44002.1
KQ459602
KPI93194.1
+ More
KQ461178 KPJ07769.1 PCG78967.1 RSAL01000008 RVE53970.1 PCG78963.1 RVE53971.1 JTDY01003263 KOB69848.1 AGBW02013713 OWR42929.1 U67866 AAB53257.1 JTDY01003650 KOB69211.1 RSAL01000068 RVE49259.1 ODYU01001294 SOQ37311.1 BABH01033637 ODYU01012325 SOQ58579.1 NWSH01000591 PCG75472.1 JTDY01005583 KOB66860.1 NWSH01003873 PCG65731.1 GEDC01024813 JAS12485.1 NNAY01001760 OXU23005.1 BABH01003987 BABH01003988 KQ434960 KZC12654.1 U67867 AAB53258.1 KZ288370 PBC26760.1 KQ435885 KOX69550.1 GL445739 EFN89136.1 GL449898 EFN81866.1 AGBW02007651 OWR55047.1 LBMM01000911 KMQ97230.1 KQ761246 OAD57941.1 GL438820 EFN68400.1 KQ971344 EFA05201.1 GECU01006695 JAT01012.1 GBYB01001820 JAG71587.1 KQ414663 KOC65238.1 KK107139 EZA57700.1 KQ976440 KYM86527.1 KQ459582 KPI99075.1 ODYU01009273 SOQ53573.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 NEVH01022635 PNF18738.1 EFA05204.2 ODYU01002245 SOQ39471.1 KQ982480 KYQ55999.1 GEZM01035401 JAV83262.1 ODYU01006431 SOQ48269.1 CVRI01000051 CRK99510.1 GL448189 EFN85168.1 DS235863 EEB19298.1 GL888175 EGI66082.1 PYGN01016841 PSN28987.1 KY797274 ATL76713.1 KB632169 ERL89446.1 AXCM01001692 KQ981954 KYN32228.1 KQ982519 KYQ55543.1 GALX01004174 JAB64292.1 KQ977574 KYN01849.1 KQ976413 KYM90403.1 MF817713 AXE71619.1 KA647365 AFP61994.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KQ977642 KYN01094.1 GL451785 EFN78551.1
KQ461178 KPJ07769.1 PCG78967.1 RSAL01000008 RVE53970.1 PCG78963.1 RVE53971.1 JTDY01003263 KOB69848.1 AGBW02013713 OWR42929.1 U67866 AAB53257.1 JTDY01003650 KOB69211.1 RSAL01000068 RVE49259.1 ODYU01001294 SOQ37311.1 BABH01033637 ODYU01012325 SOQ58579.1 NWSH01000591 PCG75472.1 JTDY01005583 KOB66860.1 NWSH01003873 PCG65731.1 GEDC01024813 JAS12485.1 NNAY01001760 OXU23005.1 BABH01003987 BABH01003988 KQ434960 KZC12654.1 U67867 AAB53258.1 KZ288370 PBC26760.1 KQ435885 KOX69550.1 GL445739 EFN89136.1 GL449898 EFN81866.1 AGBW02007651 OWR55047.1 LBMM01000911 KMQ97230.1 KQ761246 OAD57941.1 GL438820 EFN68400.1 KQ971344 EFA05201.1 GECU01006695 JAT01012.1 GBYB01001820 JAG71587.1 KQ414663 KOC65238.1 KK107139 EZA57700.1 KQ976440 KYM86527.1 KQ459582 KPI99075.1 ODYU01009273 SOQ53573.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 NEVH01022635 PNF18738.1 EFA05204.2 ODYU01002245 SOQ39471.1 KQ982480 KYQ55999.1 GEZM01035401 JAV83262.1 ODYU01006431 SOQ48269.1 CVRI01000051 CRK99510.1 GL448189 EFN85168.1 DS235863 EEB19298.1 GL888175 EGI66082.1 PYGN01016841 PSN28987.1 KY797274 ATL76713.1 KB632169 ERL89446.1 AXCM01001692 KQ981954 KYN32228.1 KQ982519 KYQ55543.1 GALX01004174 JAB64292.1 KQ977574 KYN01849.1 KQ976413 KYM90403.1 MF817713 AXE71619.1 KA647365 AFP61994.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KQ977642 KYN01094.1 GL451785 EFN78551.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000005204 UP000215335 UP000076502 UP000002358 UP000242457 UP000053105 UP000008237 UP000005203 UP000036403 UP000000311 UP000007266 UP000053825 UP000053097 UP000078540 UP000005205 UP000235965 UP000075809 UP000183832 UP000009046 UP000007755 UP000245037 UP000030742 UP000075883 UP000075920 UP000078541 UP000078542 UP000075880 UP000019118
UP000005204 UP000215335 UP000076502 UP000002358 UP000242457 UP000053105 UP000008237 UP000005203 UP000036403 UP000000311 UP000007266 UP000053825 UP000053097 UP000078540 UP000005205 UP000235965 UP000075809 UP000183832 UP000009046 UP000007755 UP000245037 UP000030742 UP000075883 UP000075920 UP000078541 UP000078542 UP000075880 UP000019118
PRIDE
Pfam
Interpro
IPR014030
Ketoacyl_synth_N
+ More
IPR020843 PKS_ER
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR013149 ADH_C
IPR018201 Ketoacyl_synth_AS
IPR013968 PKS_KR
IPR036736 ACP-like_sf
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR006162 Ppantetheine_attach_site
IPR020843 PKS_ER
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR013149 ADH_C
IPR018201 Ketoacyl_synth_AS
IPR013968 PKS_KR
IPR036736 ACP-like_sf
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR006162 Ppantetheine_attach_site
SUPFAM
ProteinModelPortal
A0A2A4K3Y0
A0A437B0L0
A0A194PKD7
A0A0N1I4Z6
A0A2A4K3Y5
A0A437BUA6
+ More
A0A2A4K5B4 A0A437BUJ8 A0A0L7L2W8 A0A212EN78 O01677 A0A0L7L1N0 A0A3S2NJM7 A0A2H1V933 H9JGF5 A0A2H1WZR1 A0A2A4JVR3 A0A0L7KUR1 A0A2A4J169 A0A1B6CGM1 A0A232EXC2 H9JU35 A0A154PMH1 O01678 K7IZC8 A0A2A3E4Z2 A0A0M8ZRC2 E2B583 A0A088ALX8 E2BR60 A0A212FMU5 A0A0J7NXT7 A0A310SRI2 E2ADN8 D2A4M7 A0A1B6JP24 A0A0C9R1I7 A0A0L7R328 A0A026WNV9 A0A195BMY6 A0A194Q6Q7 A0A2H1WKG4 A0A158NZW8 A0A2J7PQY0 D2A4M4 A0A2H1VFB6 K7IZC9 A0A151X6P9 A0A1Y1MBP7 A0A2H1W5A9 A0A1J1IKA4 E2BGH5 E0W0Z3 F4WIF1 A0A2P8XAG4 A0A2C9PHR5 U4UFT3 A0A182M3X3 A0A182VQY2 A0A195EVF8 A0A151X557 V5GRN6 A0A195CP13 A0A182J9B8 A0A151I5V4 A0A344X1T4 T1PGU1 N6TYU8 A0A195CLZ5 E2C0H8
A0A2A4K5B4 A0A437BUJ8 A0A0L7L2W8 A0A212EN78 O01677 A0A0L7L1N0 A0A3S2NJM7 A0A2H1V933 H9JGF5 A0A2H1WZR1 A0A2A4JVR3 A0A0L7KUR1 A0A2A4J169 A0A1B6CGM1 A0A232EXC2 H9JU35 A0A154PMH1 O01678 K7IZC8 A0A2A3E4Z2 A0A0M8ZRC2 E2B583 A0A088ALX8 E2BR60 A0A212FMU5 A0A0J7NXT7 A0A310SRI2 E2ADN8 D2A4M7 A0A1B6JP24 A0A0C9R1I7 A0A0L7R328 A0A026WNV9 A0A195BMY6 A0A194Q6Q7 A0A2H1WKG4 A0A158NZW8 A0A2J7PQY0 D2A4M4 A0A2H1VFB6 K7IZC9 A0A151X6P9 A0A1Y1MBP7 A0A2H1W5A9 A0A1J1IKA4 E2BGH5 E0W0Z3 F4WIF1 A0A2P8XAG4 A0A2C9PHR5 U4UFT3 A0A182M3X3 A0A182VQY2 A0A195EVF8 A0A151X557 V5GRN6 A0A195CP13 A0A182J9B8 A0A151I5V4 A0A344X1T4 T1PGU1 N6TYU8 A0A195CLZ5 E2C0H8
PDB
6FN6
E-value=4.58226e-49,
Score=496
Ontologies
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.637095
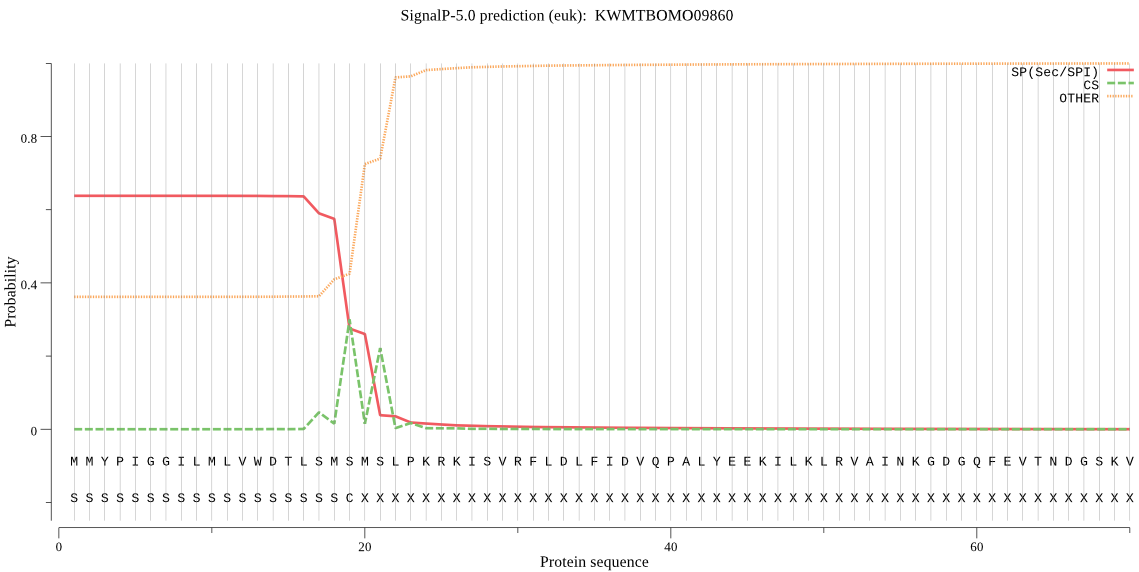
Length:
1139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.39308
Exp number, first 60 AAs:
0.16587
Total prob of N-in:
0.00910
outside
1 - 1139

Population Genetic Test Statistics
Pi
285.795209
Theta
218.712061
Tajima's D
1.225083
CLR
0.016553
CSRT
0.716314184290785
Interpretation
Uncertain
