Gene
KWMTBOMO09857
Pre Gene Modal
BGIBMGA013152
Annotation
PREDICTED:_fatty_acid_synthase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.5 Mitochondrial Reliability : 1.377 Nuclear Reliability : 1.052
Sequence
CDS
ATGGTAATAACTGATGATCATTTGAAAACCGAAATATGTTTGCCAAGAAATAATGACGGATACCCAGCTACTATACTTCACCTCGGTGCTTTGAGAAAGGACCGCGATATATCGTCTTCCCAAATTAATCGTCTGTGTGTGGAAGCTGTAGAGGCTGCCTTGTGCTCCGGAGAATATGTAGTGATTGCAGCTCCGCATGTTATTCCTCGAAAACCGCTCGTACAACAACTATGCAGTATCGCTGGTTTAGAAATGTCAGAGGATTCTAAAGACACAACGACTCTTCGCGAGCTGGGTGTCGTAGCTGAGAGAATGCCAAATATAACAAACTATTTACGGGACTATCATAACATCACGTTGATTGACAGTGATGTGTCTTCTCTAACATTGCAAAGAATTAACGAGATAGAGGACGCTATTGTTGCGCCTGGTTATAGTACTAAGAATGGGTTGGAAGCGTTCTACTCATATGTAGATCCAGACGAACTGCTAGCCACAACTGAAATGGTGTTCATGCCGACACTCACCAGTAGCGCGACCATGCGTGATGATGAGTTCGACGTGAGAGTGACATCGCTGTGTATCGTCCCGGGTCTGGAGGGACACCACGGGCGGTTCCGAGCTTTGTGCGAGCGACTCAAGCTGCCCGCGCTCGTGCTGCAACCCGGGCTGGACAAGCTGGACGAGACCATACAGCACACAGCACAACGATATGCTAGTTCTCTGCTAAGAAAATCGGCGATCAAAAATAATTTCTACATTTTGGGATATGAAACTGGTATATTTGTTGCGTTAGAACTCGCGGCTATTCTGGAAGAACACGGTATGACGGGTACATTGTTTTTGTTGGGAGGATCACCAAATGATGTGTTGAATGAGATAGAATATCAATTGCAAAGATTTAAAACAGAAAAGCTCCGAGAGGCTGTGGCAAGGCACATGCTTAAAATTATGGCCCGTGAACACTTTAGGGAAGTAGAGAACGCATTTTCGAAACGTACTCTGTGGAATGACCGAGTGGATGCGGGAGTACGAACGCTGTTGGGTCACGTGTCCCACTCGGCGCAGTACGCGCGCGCACAGATCGAGGCGGCGTGCGCGCGGCTCGTCTACGCGCGGCGCTACGGCGGCCCGGCGGCGCCCCTGCGCTCCCGGCTGGTGCTGCTGCGCTCTCCGCTCACCGCGCGCTCCGCTCCGCCAGAATGGCAGCGGCTGTCGCGGGGAGGGGTCACCGAATACAACTTGCTGGCGCCGTTACCGCATGCCACCATGGACATGCGTTGCGCTACCATCATCAACAAATATCTGGACGATGATATTTTAGACCAATTCGATAAGAAGAATTTGTGTGAAACTTACCTTCTAAATGCGGATACTTTCATGTCTGACTCAGTAATGCAGGAAAAGTAA
Protein
MVITDDHLKTEICLPRNNDGYPATILHLGALRKDRDISSSQINRLCVEAVEAALCSGEYVVIAAPHVIPRKPLVQQLCSIAGLEMSEDSKDTTTLRELGVVAERMPNITNYLRDYHNITLIDSDVSSLTLQRINEIEDAIVAPGYSTKNGLEAFYSYVDPDELLATTEMVFMPTLTSSATMRDDEFDVRVTSLCIVPGLEGHHGRFRALCERLKLPALVLQPGLDKLDETIQHTAQRYASSLLRKSAIKNNFYILGYETGIFVALELAAILEEHGMTGTLFLLGGSPNDVLNEIEYQLQRFKTEKLREAVARHMLKIMAREHFREVENAFSKRTLWNDRVDAGVRTLLGHVSHSAQYARAQIEAACARLVYARRYGGPAAPLRSRLVLLRSPLTARSAPPEWQRLSRGGVTEYNLLAPLPHATMDMRCATIINKYLDDDILDQFDKKNLCETYLLNADTFMSDSVMQEK
Summary
Uniprot
H9JUD9
H9JUS3
A0A0L7L1N0
A0A0L7L2W8
A0A0N1I4Z6
A0A194PKD7
+ More
A0A2A4K4K9 A0A2A4K3Y0 A0A2A4K4X5 A0A2A4K3Y5 A0A212EN78 A0A1L8D6D5 A0A437BSV2 A0A437B0L0 A0A0L7K4D9 H9JUE2 A0A437BUJ8 A0A3S2M8J0 A0A437BUA6 A0A2A4K5B4 O01677 A0A2H1WZR1 H9JGF5 A0A0L7L8P7 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A2H1V933 A0A0L7LMY3 A0A2A4JVR3 H9JGD5 A0A194QFD5 A0A212FMU5 H9JGD2 I4DP18 A0A1B6GFS4 A0A0L7R328 A0A310SRI2 E2ADN8 Q8MZH8 D2NUK3 G7H848 A0A1B6CGM1 Q7PLB8 A0A154PMH1 A0A1B6JP24 A0A0M8ZRC2 A0A1B6KCK1 A0A182M3X3 B5DVP9 E1ZXI7 A0A182YJT7 A0A182SD92 A0A182J9B8 A0A087ZKN3 A0A336L600 A0A1J1I2M2 A0A2A3E4Z2 A0A182R1J1 A0A182JQR8 A0A084VRX8 B3N4Q4 B4LXF7 A0A0C9R1I7 E2C0H8 A0A1W4VE27
A0A2A4K4K9 A0A2A4K3Y0 A0A2A4K4X5 A0A2A4K3Y5 A0A212EN78 A0A1L8D6D5 A0A437BSV2 A0A437B0L0 A0A0L7K4D9 H9JUE2 A0A437BUJ8 A0A3S2M8J0 A0A437BUA6 A0A2A4K5B4 O01677 A0A2H1WZR1 H9JGF5 A0A0L7L8P7 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A2H1V933 A0A0L7LMY3 A0A2A4JVR3 H9JGD5 A0A194QFD5 A0A212FMU5 H9JGD2 I4DP18 A0A1B6GFS4 A0A0L7R328 A0A310SRI2 E2ADN8 Q8MZH8 D2NUK3 G7H848 A0A1B6CGM1 Q7PLB8 A0A154PMH1 A0A1B6JP24 A0A0M8ZRC2 A0A1B6KCK1 A0A182M3X3 B5DVP9 E1ZXI7 A0A182YJT7 A0A182SD92 A0A182J9B8 A0A087ZKN3 A0A336L600 A0A1J1I2M2 A0A2A3E4Z2 A0A182R1J1 A0A182JQR8 A0A084VRX8 B3N4Q4 B4LXF7 A0A0C9R1I7 E2C0H8 A0A1W4VE27
Pubmed
EMBL
BABH01022901
BABH01022902
BABH01041756
JTDY01003650
KOB69211.1
JTDY01003263
+ More
KOB69848.1 KQ461178 KPJ07769.1 KQ459602 KPI93194.1 NWSH01000156 PCG78966.1 PCG78965.1 PCG78964.1 PCG78967.1 AGBW02013713 OWR42929.1 GEYN01000117 JAV02012.1 RSAL01000013 RVE53441.1 RSAL01000223 RVE44002.1 JTDY01012055 KOB52337.1 BABH01022891 RSAL01000008 RVE53971.1 RVE53445.1 RVE53970.1 PCG78963.1 U67866 AAB53257.1 ODYU01012325 SOQ58579.1 BABH01033637 JTDY01002311 KOB71689.1 NWSH01000591 PCG75470.1 RSAL01000068 RVE49259.1 PCG75471.1 ODYU01001294 SOQ37311.1 JTDY01000529 KOB76790.1 PCG75472.1 BABH01033612 BABH01033613 KQ459053 KPJ04124.1 AGBW02007651 OWR55047.1 BABH01033622 BABH01033623 BABH01033624 AK403251 BAM19658.1 GECZ01008485 JAS61284.1 KQ414663 KOC65238.1 KQ761246 OAD57941.1 GL438820 EFN68400.1 AY102675 AAM27504.1 BT120146 ADB57067.1 BT132772 AET14856.1 GEDC01024813 JAS12485.1 AE014296 EAA46042.3 KQ434960 KZC12654.1 GECU01006695 JAT01012.1 KQ435885 KOX69550.1 GEBQ01031053 JAT08924.1 AXCM01001692 CM000070 EDY68457.3 GL435066 EFN74070.1 UFQS01001117 UFQT01001117 SSX09070.1 SSX28981.1 CVRI01000038 CRK94451.1 KZ288370 PBC26760.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 CH954177 EDV60008.2 CH940650 EDW67835.2 GBYB01001820 JAG71587.1 GL451785 EFN78551.1
KOB69848.1 KQ461178 KPJ07769.1 KQ459602 KPI93194.1 NWSH01000156 PCG78966.1 PCG78965.1 PCG78964.1 PCG78967.1 AGBW02013713 OWR42929.1 GEYN01000117 JAV02012.1 RSAL01000013 RVE53441.1 RSAL01000223 RVE44002.1 JTDY01012055 KOB52337.1 BABH01022891 RSAL01000008 RVE53971.1 RVE53445.1 RVE53970.1 PCG78963.1 U67866 AAB53257.1 ODYU01012325 SOQ58579.1 BABH01033637 JTDY01002311 KOB71689.1 NWSH01000591 PCG75470.1 RSAL01000068 RVE49259.1 PCG75471.1 ODYU01001294 SOQ37311.1 JTDY01000529 KOB76790.1 PCG75472.1 BABH01033612 BABH01033613 KQ459053 KPJ04124.1 AGBW02007651 OWR55047.1 BABH01033622 BABH01033623 BABH01033624 AK403251 BAM19658.1 GECZ01008485 JAS61284.1 KQ414663 KOC65238.1 KQ761246 OAD57941.1 GL438820 EFN68400.1 AY102675 AAM27504.1 BT120146 ADB57067.1 BT132772 AET14856.1 GEDC01024813 JAS12485.1 AE014296 EAA46042.3 KQ434960 KZC12654.1 GECU01006695 JAT01012.1 KQ435885 KOX69550.1 GEBQ01031053 JAT08924.1 AXCM01001692 CM000070 EDY68457.3 GL435066 EFN74070.1 UFQS01001117 UFQT01001117 SSX09070.1 SSX28981.1 CVRI01000038 CRK94451.1 KZ288370 PBC26760.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 CH954177 EDV60008.2 CH940650 EDW67835.2 GBYB01001820 JAG71587.1 GL451785 EFN78551.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000283053 UP000053825 UP000000311 UP000000803 UP000076502 UP000053105 UP000075883 UP000001819 UP000076408 UP000075901 UP000075880 UP000183832 UP000242457 UP000075900 UP000075881 UP000030765 UP000008711 UP000008792 UP000008237 UP000192221
UP000283053 UP000053825 UP000000311 UP000000803 UP000076502 UP000053105 UP000075883 UP000001819 UP000076408 UP000075901 UP000075880 UP000183832 UP000242457 UP000075900 UP000075881 UP000030765 UP000008711 UP000008792 UP000008237 UP000192221
Pfam
Interpro
IPR023102
Fatty_acid_synthase_dom_2
+ More
IPR029058 AB_hydrolase
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR011032 GroES-like_sf
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR029058 AB_hydrolase
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR011032 GroES-like_sf
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
SUPFAM
CDD
ProteinModelPortal
H9JUD9
H9JUS3
A0A0L7L1N0
A0A0L7L2W8
A0A0N1I4Z6
A0A194PKD7
+ More
A0A2A4K4K9 A0A2A4K3Y0 A0A2A4K4X5 A0A2A4K3Y5 A0A212EN78 A0A1L8D6D5 A0A437BSV2 A0A437B0L0 A0A0L7K4D9 H9JUE2 A0A437BUJ8 A0A3S2M8J0 A0A437BUA6 A0A2A4K5B4 O01677 A0A2H1WZR1 H9JGF5 A0A0L7L8P7 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A2H1V933 A0A0L7LMY3 A0A2A4JVR3 H9JGD5 A0A194QFD5 A0A212FMU5 H9JGD2 I4DP18 A0A1B6GFS4 A0A0L7R328 A0A310SRI2 E2ADN8 Q8MZH8 D2NUK3 G7H848 A0A1B6CGM1 Q7PLB8 A0A154PMH1 A0A1B6JP24 A0A0M8ZRC2 A0A1B6KCK1 A0A182M3X3 B5DVP9 E1ZXI7 A0A182YJT7 A0A182SD92 A0A182J9B8 A0A087ZKN3 A0A336L600 A0A1J1I2M2 A0A2A3E4Z2 A0A182R1J1 A0A182JQR8 A0A084VRX8 B3N4Q4 B4LXF7 A0A0C9R1I7 E2C0H8 A0A1W4VE27
A0A2A4K4K9 A0A2A4K3Y0 A0A2A4K4X5 A0A2A4K3Y5 A0A212EN78 A0A1L8D6D5 A0A437BSV2 A0A437B0L0 A0A0L7K4D9 H9JUE2 A0A437BUJ8 A0A3S2M8J0 A0A437BUA6 A0A2A4K5B4 O01677 A0A2H1WZR1 H9JGF5 A0A0L7L8P7 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A2H1V933 A0A0L7LMY3 A0A2A4JVR3 H9JGD5 A0A194QFD5 A0A212FMU5 H9JGD2 I4DP18 A0A1B6GFS4 A0A0L7R328 A0A310SRI2 E2ADN8 Q8MZH8 D2NUK3 G7H848 A0A1B6CGM1 Q7PLB8 A0A154PMH1 A0A1B6JP24 A0A0M8ZRC2 A0A1B6KCK1 A0A182M3X3 B5DVP9 E1ZXI7 A0A182YJT7 A0A182SD92 A0A182J9B8 A0A087ZKN3 A0A336L600 A0A1J1I2M2 A0A2A3E4Z2 A0A182R1J1 A0A182JQR8 A0A084VRX8 B3N4Q4 B4LXF7 A0A0C9R1I7 E2C0H8 A0A1W4VE27
PDB
2VZ9
E-value=8.00466e-06,
Score=119
Ontologies
KEGG
PATHWAY
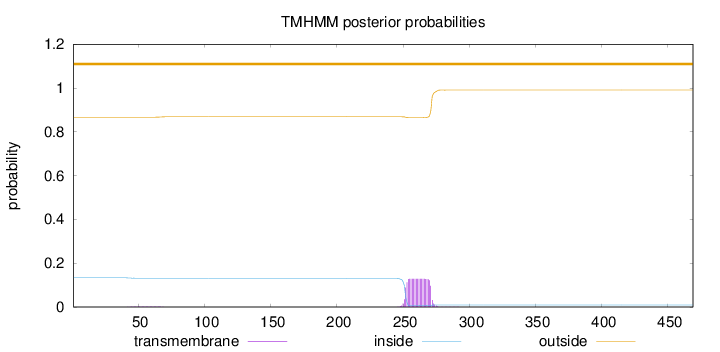
Topology
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.64869
Exp number, first 60 AAs:
0.03164
Total prob of N-in:
0.13319
outside
1 - 469
Population Genetic Test Statistics
Pi
199.887838
Theta
166.763678
Tajima's D
0.606959
CLR
0.008646
CSRT
0.547522623868807
Interpretation
Uncertain
