Gene
KWMTBOMO09848
Pre Gene Modal
BGIBMGA013280
Annotation
PREDICTED:_uncharacterized_protein_LOC105841877?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.122
Sequence
CDS
ATGACCACTCTACAAGATTTGGCTGCTGGAAGTGACAATATAAAATTAATTAAGGAATTTTTACGTCTGAAATATCACGTTTGTATTTCGGAACACAACGTTTTGAAGTTAAATAAACAAAAACTTCGTGAGATAGATTTTCTTGCAAACAACTTTGAGCACAAACAAGGTTTGGCTGTTTACTTCTCTCACATAGCTGAAGATGAACTCAGCTCCGATACTCCTATATATTTATTGCCTACTCTTGGTAATGCAAATGCGGAAAAACAGGTCGATGCACTGAAGTCTGAGTACTTGTGCGTTGTTCCTGGGATTGAAGGTCATCACGTAAGATTTCGTGGTCTCTGTGAGAGACTCAAACTGCCCGCGCTGGTGCTGCAGCCAAGTTTGAATGTGTTGCACGAAACCATAGAACAGGCTGCTCAAAGCTACTCTAAGATACTCATGGAAAAAATTGGTGTCGGTAACAATTTCTACTTGCTTGGGTGTGATATTGGAATTTTGATTGCGATGGAACTAGCTGCAATACTTGAGAGAAATGGTTTTTTGGGCACCATATTTTGTCTCGGAACTACACCAGAAGACGTCAAGCCAGTAATTGAACAACAACTAGGGGCGTTCTCAACAGAAGACCAACTGCAGCTTGGAATTTTAAAACATATTTTTACGCTACTAACTGGTGAAGTACCATCAGATTTAGATGAAACTCTGGACCCTCACTCTTCATGGAATGAAAAAATTGACGCTTGTATCAAAATGATGGCCGGTCGTATAACACAAACAAAGCAATATGTGAAAGCGTTAATAAATTTCGTCTATGAAAGAATTCTTAGCATCAAACAGTATGAATCCTCATCTTTGCCAATACTGAAGTCTAGAGTCGTGATACTCCGCTCCCCAGGATATTCGTCAACTTCATCTGCATCATCATACGTAATTGAAGATATATCAGCACCACTTTGTGATGCTGCAAAAGATGTGTGA
Protein
MTTLQDLAAGSDNIKLIKEFLRLKYHVCISEHNVLKLNKQKLREIDFLANNFEHKQGLAVYFSHIAEDELSSDTPIYLLPTLGNANAEKQVDALKSEYLCVVPGIEGHHVRFRGLCERLKLPALVLQPSLNVLHETIEQAAQSYSKILMEKIGVGNNFYLLGCDIGILIAMELAAILERNGFLGTIFCLGTTPEDVKPVIEQQLGAFSTEDQLQLGILKHIFTLLTGEVPSDLDETLDPHSSWNEKIDACIKMMAGRITQTKQYVKALINFVYERILSIKQYESSSLPILKSRVVILRSPGYSSTSSASSYVIEDISAPLCDAAKDV
Summary
Uniprot
H9JUR7
H9JUE2
A0A2A4K4X5
A0A0L7L1N0
H9JUS3
A0A2A4K3Y5
+ More
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y0 H9JUD9 A0A0L7K4D9 A0A0F6Q2Z3 A0A0L7L2W8 A0A194PKD7 A0A437BSV2 A0A437B0L0 A0A212EN78 A0A0L7L2S0 A0A0L7LPX5 A0A0L7L594 A0A1L8D6D5 A0A3S2M8J0 A0A437BUA6 A0A3S2NJM7 A0A2A4K5B4 A0A2H1WZR1 A0A2A4JTZ4 O01677 A0A2A4JVQ1 A0A437BUJ8 H9JGF5 A0A2H1V933 A0A0L7L8P7 A0A0L7LMK6 A0A0L7LMY3 H9JGD5 A0A2A4JVR3 A0A194QF64 H9JU35 O01678 A0A194QF04 A0A2A4J169 A0A2J7PFX6 A0A2J7PFX2 A0A2H1WSB0 E2ADN8 A0A212FMU5 E2C0H8 K7IZC8 A0A2H1VFB6 J9K3D3 A0A1B6CGM1 A0A232EXC2 A0A0J7MTJ2 A0A1J1IGX7 E9ITY4 A0A2S2NJ15 A0A0C9R0U2 A0A0G3YL52 A0A1B6GFS4 A0A310SRI2 E1ZXI7 A0A232EX94 A0A2J7PQY0 E2BR60 A0A1B6KCK1 A0A195BMY6 A0A151X6P9 A0A2S2RA89 A0A1B6JP24 I4DP18 A0A158NZW8 F4WIF1 A0A0C9R1I7 E2B583 B0WMC9 A0A1B0CP42 E2ABR4
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y0 H9JUD9 A0A0L7K4D9 A0A0F6Q2Z3 A0A0L7L2W8 A0A194PKD7 A0A437BSV2 A0A437B0L0 A0A212EN78 A0A0L7L2S0 A0A0L7LPX5 A0A0L7L594 A0A1L8D6D5 A0A3S2M8J0 A0A437BUA6 A0A3S2NJM7 A0A2A4K5B4 A0A2H1WZR1 A0A2A4JTZ4 O01677 A0A2A4JVQ1 A0A437BUJ8 H9JGF5 A0A2H1V933 A0A0L7L8P7 A0A0L7LMK6 A0A0L7LMY3 H9JGD5 A0A2A4JVR3 A0A194QF64 H9JU35 O01678 A0A194QF04 A0A2A4J169 A0A2J7PFX6 A0A2J7PFX2 A0A2H1WSB0 E2ADN8 A0A212FMU5 E2C0H8 K7IZC8 A0A2H1VFB6 J9K3D3 A0A1B6CGM1 A0A232EXC2 A0A0J7MTJ2 A0A1J1IGX7 E9ITY4 A0A2S2NJ15 A0A0C9R0U2 A0A0G3YL52 A0A1B6GFS4 A0A310SRI2 E1ZXI7 A0A232EX94 A0A2J7PQY0 E2BR60 A0A1B6KCK1 A0A195BMY6 A0A151X6P9 A0A2S2RA89 A0A1B6JP24 I4DP18 A0A158NZW8 F4WIF1 A0A0C9R1I7 E2B583 B0WMC9 A0A1B0CP42 E2ABR4
Pubmed
EMBL
BABH01022926
BABH01022927
BABH01022928
BABH01022891
NWSH01000156
PCG78964.1
+ More
JTDY01003650 KOB69211.1 BABH01041756 PCG78967.1 PCG78966.1 KQ461178 KPJ07769.1 PCG78965.1 BABH01022901 BABH01022902 JTDY01012055 KOB52337.1 KP237907 AKD01760.1 JTDY01003263 KOB69848.1 KQ459602 KPI93194.1 RSAL01000013 RVE53441.1 RSAL01000223 RVE44002.1 AGBW02013713 OWR42929.1 JTDY01003335 KOB69710.1 JTDY01000426 KOB77241.1 JTDY01002859 KOB70590.1 GEYN01000117 JAV02012.1 RVE53445.1 RSAL01000008 RVE53970.1 RSAL01000068 RVE49259.1 PCG78963.1 ODYU01012325 SOQ58579.1 NWSH01000591 PCG75471.1 U67866 AAB53257.1 PCG75470.1 RVE53971.1 BABH01033637 ODYU01001294 SOQ37311.1 JTDY01002311 KOB71689.1 JTDY01000529 KOB76788.1 KOB76790.1 BABH01033612 BABH01033613 PCG75472.1 KQ459053 KPJ04122.1 BABH01003987 BABH01003988 U67867 AAB53258.1 KPJ04123.1 NWSH01003873 PCG65731.1 NEVH01025646 PNF15228.1 PNF15226.1 ODYU01010623 SOQ55856.1 GL438820 EFN68400.1 AGBW02007651 OWR55047.1 GL451785 EFN78551.1 ODYU01002245 SOQ39471.1 ABLF02036897 GEDC01024813 JAS12485.1 NNAY01001760 OXU23005.1 LBMM01018330 KMQ83820.1 CVRI01000051 CRK99509.1 GL765755 EFZ15969.1 GGMR01004548 MBY17167.1 GBYB01001605 JAG71372.1 KM507100 AKM28424.1 GECZ01008485 JAS61284.1 KQ761246 OAD57941.1 GL435066 EFN74070.1 OXU23002.1 NEVH01022635 PNF18738.1 GL449898 EFN81866.1 GEBQ01031053 JAT08924.1 KQ976440 KYM86527.1 KQ982480 KYQ55999.1 GGMS01017661 MBY86864.1 GECU01006695 JAT01012.1 AK403251 BAM19658.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 GL888175 EGI66082.1 GBYB01001820 JAG71587.1 GL445739 EFN89136.1 DS231997 EDS30986.1 AJWK01021412 GL438338 EFN69128.1
JTDY01003650 KOB69211.1 BABH01041756 PCG78967.1 PCG78966.1 KQ461178 KPJ07769.1 PCG78965.1 BABH01022901 BABH01022902 JTDY01012055 KOB52337.1 KP237907 AKD01760.1 JTDY01003263 KOB69848.1 KQ459602 KPI93194.1 RSAL01000013 RVE53441.1 RSAL01000223 RVE44002.1 AGBW02013713 OWR42929.1 JTDY01003335 KOB69710.1 JTDY01000426 KOB77241.1 JTDY01002859 KOB70590.1 GEYN01000117 JAV02012.1 RVE53445.1 RSAL01000008 RVE53970.1 RSAL01000068 RVE49259.1 PCG78963.1 ODYU01012325 SOQ58579.1 NWSH01000591 PCG75471.1 U67866 AAB53257.1 PCG75470.1 RVE53971.1 BABH01033637 ODYU01001294 SOQ37311.1 JTDY01002311 KOB71689.1 JTDY01000529 KOB76788.1 KOB76790.1 BABH01033612 BABH01033613 PCG75472.1 KQ459053 KPJ04122.1 BABH01003987 BABH01003988 U67867 AAB53258.1 KPJ04123.1 NWSH01003873 PCG65731.1 NEVH01025646 PNF15228.1 PNF15226.1 ODYU01010623 SOQ55856.1 GL438820 EFN68400.1 AGBW02007651 OWR55047.1 GL451785 EFN78551.1 ODYU01002245 SOQ39471.1 ABLF02036897 GEDC01024813 JAS12485.1 NNAY01001760 OXU23005.1 LBMM01018330 KMQ83820.1 CVRI01000051 CRK99509.1 GL765755 EFZ15969.1 GGMR01004548 MBY17167.1 GBYB01001605 JAG71372.1 KM507100 AKM28424.1 GECZ01008485 JAS61284.1 KQ761246 OAD57941.1 GL435066 EFN74070.1 OXU23002.1 NEVH01022635 PNF18738.1 GL449898 EFN81866.1 GEBQ01031053 JAT08924.1 KQ976440 KYM86527.1 KQ982480 KYQ55999.1 GGMS01017661 MBY86864.1 GECU01006695 JAT01012.1 AK403251 BAM19658.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 GL888175 EGI66082.1 GBYB01001820 JAG71587.1 GL445739 EFN89136.1 DS231997 EDS30986.1 AJWK01021412 GL438338 EFN69128.1
Proteomes
Pfam
Interpro
IPR029058
AB_hydrolase
+ More
IPR042104 PKS_dehydratase_sf
IPR023102 Fatty_acid_synthase_dom_2
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR011032 GroES-like_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR020807 PKS_dehydratase
IPR013968 PKS_KR
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR006162 Ppantetheine_attach_site
IPR000999 RNase_III_dom
IPR036389 RNase_III_sf
IPR042104 PKS_dehydratase_sf
IPR023102 Fatty_acid_synthase_dom_2
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR011032 GroES-like_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR020807 PKS_dehydratase
IPR013968 PKS_KR
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR006162 Ppantetheine_attach_site
IPR000999 RNase_III_dom
IPR036389 RNase_III_sf
SUPFAM
CDD
ProteinModelPortal
H9JUR7
H9JUE2
A0A2A4K4X5
A0A0L7L1N0
H9JUS3
A0A2A4K3Y5
+ More
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y0 H9JUD9 A0A0L7K4D9 A0A0F6Q2Z3 A0A0L7L2W8 A0A194PKD7 A0A437BSV2 A0A437B0L0 A0A212EN78 A0A0L7L2S0 A0A0L7LPX5 A0A0L7L594 A0A1L8D6D5 A0A3S2M8J0 A0A437BUA6 A0A3S2NJM7 A0A2A4K5B4 A0A2H1WZR1 A0A2A4JTZ4 O01677 A0A2A4JVQ1 A0A437BUJ8 H9JGF5 A0A2H1V933 A0A0L7L8P7 A0A0L7LMK6 A0A0L7LMY3 H9JGD5 A0A2A4JVR3 A0A194QF64 H9JU35 O01678 A0A194QF04 A0A2A4J169 A0A2J7PFX6 A0A2J7PFX2 A0A2H1WSB0 E2ADN8 A0A212FMU5 E2C0H8 K7IZC8 A0A2H1VFB6 J9K3D3 A0A1B6CGM1 A0A232EXC2 A0A0J7MTJ2 A0A1J1IGX7 E9ITY4 A0A2S2NJ15 A0A0C9R0U2 A0A0G3YL52 A0A1B6GFS4 A0A310SRI2 E1ZXI7 A0A232EX94 A0A2J7PQY0 E2BR60 A0A1B6KCK1 A0A195BMY6 A0A151X6P9 A0A2S2RA89 A0A1B6JP24 I4DP18 A0A158NZW8 F4WIF1 A0A0C9R1I7 E2B583 B0WMC9 A0A1B0CP42 E2ABR4
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y0 H9JUD9 A0A0L7K4D9 A0A0F6Q2Z3 A0A0L7L2W8 A0A194PKD7 A0A437BSV2 A0A437B0L0 A0A212EN78 A0A0L7L2S0 A0A0L7LPX5 A0A0L7L594 A0A1L8D6D5 A0A3S2M8J0 A0A437BUA6 A0A3S2NJM7 A0A2A4K5B4 A0A2H1WZR1 A0A2A4JTZ4 O01677 A0A2A4JVQ1 A0A437BUJ8 H9JGF5 A0A2H1V933 A0A0L7L8P7 A0A0L7LMK6 A0A0L7LMY3 H9JGD5 A0A2A4JVR3 A0A194QF64 H9JU35 O01678 A0A194QF04 A0A2A4J169 A0A2J7PFX6 A0A2J7PFX2 A0A2H1WSB0 E2ADN8 A0A212FMU5 E2C0H8 K7IZC8 A0A2H1VFB6 J9K3D3 A0A1B6CGM1 A0A232EXC2 A0A0J7MTJ2 A0A1J1IGX7 E9ITY4 A0A2S2NJ15 A0A0C9R0U2 A0A0G3YL52 A0A1B6GFS4 A0A310SRI2 E1ZXI7 A0A232EX94 A0A2J7PQY0 E2BR60 A0A1B6KCK1 A0A195BMY6 A0A151X6P9 A0A2S2RA89 A0A1B6JP24 I4DP18 A0A158NZW8 F4WIF1 A0A0C9R1I7 E2B583 B0WMC9 A0A1B0CP42 E2ABR4
Ontologies
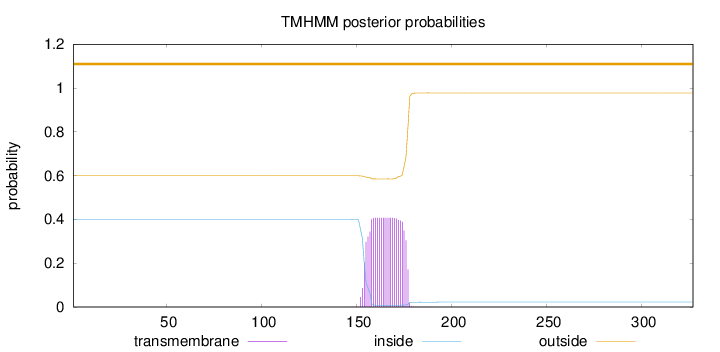
Topology
Length:
327
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.02036999999997
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.40065
outside
1 - 327
Population Genetic Test Statistics
Pi
115.545845
Theta
166.663522
Tajima's D
-1.001776
CLR
243.892261
CSRT
0.140242987850607
Interpretation
Uncertain
