Gene
KWMTBOMO09847
Pre Gene Modal
BGIBMGA013286
Annotation
PREDICTED:_fatty_acid_synthase-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.41 Nuclear Reliability : 1.285
Sequence
CDS
ATGGAGGACGCTATTGTTGCGCCTGGTTATAGTACAAAGAAAGGGTTGGAAGCGTTCTATTCGTATGTAGATCCAGACGAACTGCTAGCCACAAGTGAAATGGTGCTCATGCCGACACTCACTAGTAGTGCAACTGCGCGCGATGATGAGTTCGACGTGACTGTGACATCGCTGTGTATCGTCCCGGGTTTGGAGGGACACCATGGGCGGTTTCGAGCTTTGTGCGAGCGACTCAAGCTGCCCGCGCTCGTGCTGCAACCCGGGCTGGACAAGCTGGACGAGACCATACAGCAGACAGCACAACGATATGCTAGTTCTCTGCTAAGAAAATCAGCGATAAAAAATAATTTCTACATTTTGGGATATGAAACTGGTATATTTGTTGCGTTAGAACTCACGGCTATTCTGGAAGAACACGATATGACGGGTACGTTGTTTTTGTTGGGAGGATCACCAAATGATGTGTCGAATGAAATAGAATATCAACTGCAAGGATTCAAAACAGATCAGCTCCAGGAGGCTGTGGCAAGGCACATGTTTAAAATTATGGCCCGTGAACACTTTAAGGAAGTAGAGAACGCATTTTCGAAACGTACTCTGTGGAATGACCGAGTGGATGCGGGAGTGCGAACCCTGTTGGGTCACGTGTCCCACTCGGCGCAGTACGCGCGCGCACAGATCGAGGCGGCGTGCGCGCGGCTCGTCTACGCGCGGCGCTACGGCGGCCCGGCGGCGCCCCTACGCTCCCGGCTCGTGCTGCTGCGCTCTCCGCTCACCGCGCGCTCCGCTCCGCCAGAATGGCAGCGGCTGTCGCGGGGAGGGGTCACCGAACACAACTTGCTGGCGCCGTTACCGCATGCCGCCATGGACATGCGTTGCGCTACCATCATCAACAAATATCTGGACGATGAAATTTTAGACCAATTCAATAAGAAGAATTTGTGTGAAACGTATTTACTTAGTGCGGATACGTACATGTCTTTTGTCACTGATTTGAATTAG
Protein
MEDAIVAPGYSTKKGLEAFYSYVDPDELLATSEMVLMPTLTSSATARDDEFDVTVTSLCIVPGLEGHHGRFRALCERLKLPALVLQPGLDKLDETIQQTAQRYASSLLRKSAIKNNFYILGYETGIFVALELTAILEEHDMTGTLFLLGGSPNDVSNEIEYQLQGFKTDQLQEAVARHMFKIMAREHFKEVENAFSKRTLWNDRVDAGVRTLLGHVSHSAQYARAQIEAACARLVYARRYGGPAAPLRSRLVLLRSPLTARSAPPEWQRLSRGGVTEHNLLAPLPHAAMDMRCATIINKYLDDEILDQFNKKNLCETYLLSADTYMSFVTDLN
Summary
Uniprot
H9JUS3
H9JUD9
A0A0L7L1N0
A0A2A4K4X5
A0A0L7L2W8
A0A194PKD7
+ More
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y5 A0A2A4K3Y0 A0A212EN78 A0A437BSV2 A0A1L8D6D5 A0A437B0L0 A0A0L7LPX5 A0A0L7L594 H9JUE2 A0A0L7KUW1 A0A0L7L2E8 A0A0L7L2S0 A0A2A4K5B4 A0A437BUJ8 A0A437BUA6 A0A2H1WZR1 H9JGF5 O01677 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A194QW12 A0A0L7LMK6 A0A2A4JVR3 A0A2H1V933 A0A0L7LMY3 A0A194QF64 A0A194QF04 A0A194QFD5 H9JGD5 A0A0L7L080 A0A310SRI2 H9JGD2 I4DP18 A0A2J7PFX6 A0A194Q6Q7 A0A2J7PFX2 A0A088ALX8
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y5 A0A2A4K3Y0 A0A212EN78 A0A437BSV2 A0A1L8D6D5 A0A437B0L0 A0A0L7LPX5 A0A0L7L594 H9JUE2 A0A0L7KUW1 A0A0L7L2E8 A0A0L7L2S0 A0A2A4K5B4 A0A437BUJ8 A0A437BUA6 A0A2H1WZR1 H9JGF5 O01677 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A194QW12 A0A0L7LMK6 A0A2A4JVR3 A0A2H1V933 A0A0L7LMY3 A0A194QF64 A0A194QF04 A0A194QFD5 H9JGD5 A0A0L7L080 A0A310SRI2 H9JGD2 I4DP18 A0A2J7PFX6 A0A194Q6Q7 A0A2J7PFX2 A0A088ALX8
EMBL
BABH01041756
BABH01022901
BABH01022902
JTDY01003650
KOB69211.1
NWSH01000156
+ More
PCG78964.1 JTDY01003263 KOB69848.1 KQ459602 KPI93194.1 PCG78966.1 KQ461178 KPJ07769.1 PCG78967.1 PCG78965.1 AGBW02013713 OWR42929.1 RSAL01000013 RVE53441.1 GEYN01000117 JAV02012.1 RSAL01000223 RVE44002.1 JTDY01000426 KOB77241.1 JTDY01002859 KOB70590.1 BABH01022891 JTDY01005583 KOB66861.1 JTDY01003390 KOB69632.1 JTDY01003335 KOB69710.1 PCG78963.1 RSAL01000008 RVE53971.1 RVE53970.1 ODYU01012325 SOQ58579.1 BABH01033637 U67866 AAB53257.1 NWSH01000591 PCG75470.1 RSAL01000068 RVE49259.1 PCG75471.1 KQ461073 KPJ09499.1 JTDY01000529 KOB76788.1 PCG75472.1 ODYU01001294 SOQ37311.1 KOB76790.1 KQ459053 KPJ04122.1 KPJ04123.1 KPJ04124.1 BABH01033612 BABH01033613 JTDY01004048 KOB68669.1 KQ761246 OAD57941.1 BABH01033622 BABH01033623 BABH01033624 AK403251 BAM19658.1 NEVH01025646 PNF15228.1 KQ459582 KPI99075.1 PNF15226.1
PCG78964.1 JTDY01003263 KOB69848.1 KQ459602 KPI93194.1 PCG78966.1 KQ461178 KPJ07769.1 PCG78967.1 PCG78965.1 AGBW02013713 OWR42929.1 RSAL01000013 RVE53441.1 GEYN01000117 JAV02012.1 RSAL01000223 RVE44002.1 JTDY01000426 KOB77241.1 JTDY01002859 KOB70590.1 BABH01022891 JTDY01005583 KOB66861.1 JTDY01003390 KOB69632.1 JTDY01003335 KOB69710.1 PCG78963.1 RSAL01000008 RVE53971.1 RVE53970.1 ODYU01012325 SOQ58579.1 BABH01033637 U67866 AAB53257.1 NWSH01000591 PCG75470.1 RSAL01000068 RVE49259.1 PCG75471.1 KQ461073 KPJ09499.1 JTDY01000529 KOB76788.1 PCG75472.1 ODYU01001294 SOQ37311.1 KOB76790.1 KQ459053 KPJ04122.1 KPJ04123.1 KPJ04124.1 BABH01033612 BABH01033613 JTDY01004048 KOB68669.1 KQ761246 OAD57941.1 BABH01033622 BABH01033623 BABH01033624 AK403251 BAM19658.1 NEVH01025646 PNF15228.1 KQ459582 KPI99075.1 PNF15226.1
Proteomes
Pfam
Interpro
IPR023102
Fatty_acid_synthase_dom_2
+ More
IPR029058 AB_hydrolase
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR011032 GroES-like_sf
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR029058 AB_hydrolase
IPR042104 PKS_dehydratase_sf
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR011032 GroES-like_sf
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR018201 Ketoacyl_synth_AS
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
SUPFAM
CDD
ProteinModelPortal
H9JUS3
H9JUD9
A0A0L7L1N0
A0A2A4K4X5
A0A0L7L2W8
A0A194PKD7
+ More
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y5 A0A2A4K3Y0 A0A212EN78 A0A437BSV2 A0A1L8D6D5 A0A437B0L0 A0A0L7LPX5 A0A0L7L594 H9JUE2 A0A0L7KUW1 A0A0L7L2E8 A0A0L7L2S0 A0A2A4K5B4 A0A437BUJ8 A0A437BUA6 A0A2H1WZR1 H9JGF5 O01677 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A194QW12 A0A0L7LMK6 A0A2A4JVR3 A0A2H1V933 A0A0L7LMY3 A0A194QF64 A0A194QF04 A0A194QFD5 H9JGD5 A0A0L7L080 A0A310SRI2 H9JGD2 I4DP18 A0A2J7PFX6 A0A194Q6Q7 A0A2J7PFX2 A0A088ALX8
A0A2A4K4K9 A0A0N1I4Z6 A0A2A4K3Y5 A0A2A4K3Y0 A0A212EN78 A0A437BSV2 A0A1L8D6D5 A0A437B0L0 A0A0L7LPX5 A0A0L7L594 H9JUE2 A0A0L7KUW1 A0A0L7L2E8 A0A0L7L2S0 A0A2A4K5B4 A0A437BUJ8 A0A437BUA6 A0A2H1WZR1 H9JGF5 O01677 A0A2A4JVQ1 A0A3S2NJM7 A0A2A4JTZ4 A0A194QW12 A0A0L7LMK6 A0A2A4JVR3 A0A2H1V933 A0A0L7LMY3 A0A194QF64 A0A194QF04 A0A194QFD5 H9JGD5 A0A0L7L080 A0A310SRI2 H9JGD2 I4DP18 A0A2J7PFX6 A0A194Q6Q7 A0A2J7PFX2 A0A088ALX8
Ontologies
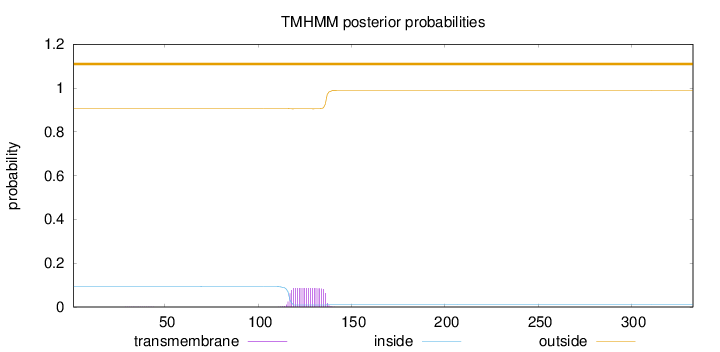
Topology
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.73897
Exp number, first 60 AAs:
0.00332
Total prob of N-in:
0.09314
outside
1 - 333
Population Genetic Test Statistics
Pi
174.496331
Theta
180.307677
Tajima's D
-0.673978
CLR
0
CSRT
0.203389830508475
Interpretation
Uncertain
