Gene
KWMTBOMO09846
Pre Gene Modal
BGIBMGA013285
Annotation
Uncharacterized_protein_OBRU01_10567?_partial_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.967 Nuclear Reliability : 2.039
Sequence
CDS
ATGAGTGAACATAGTAAAGTTGCAACAGGTTTCATAATAAAAACCAAAGTTGTAAATCAATGTATCAATGAAGTTGATTCGGATCATAAGGACATGGAATTGAACGCGGATGACATTTATCATCTACTAAAACTGCGCGACTACTCTTATAGCGGAGATTTTCGGAGTGTACATAATGGAAATTCTATACTGAATTCATTTTCAATTATTTGGAATGATAACTGGGTGACATTTATTGATGGTTTGCTACAAGTTAATGCATTAAGACAACGACATGACGGAGTATCTCAGCCCGCGGAAATTAGAAAATTATGTATAGATGTTACCGAGCACAGAAAGCATATTACGAAAGTTGATAACAAGGAAATAATACCAGCCAAGGTTCTTGAAAAGCATGAAACCACAAAAGCTGGAGGTGTATTATTGCAGGGACTATTATTCCAGCAATTACCACCGGTGATCCTTAAGGGCATCTGCCTGAAATCTTCAAAATTCATCCCTCGCTTCAACGCGAAATTAGATGATGTCTCAGCTATACATGTTTTTCTACAAATCGTAATGGAAAATATTGACAAAGACGAACTCAACGTTCTTGGAATTTTAAATGACAGCGGTCTTGTTCATCCAGCCCTCGCTAAAATCACAAATGAATTTAATGGAATATCTATACATTACAAAGAACTCAGCCGTAAGATTTTAATTACAAAAAAATCCGAACATTTAGCTGATGCAGACCTTGTAGTTGTTCGAGGCATGACCACAGATAGCTGTGGTGCATTTGGCGGTTACTACTATTTACCACTGAAACAAGAGGCTACGAAAGAGGATTGCGAATTAAAATTGGAATGTTCGGTTACTGGAAATCTTAATTCCTTGAAATGGGTGCAAAATTCTCAAAGCAGTACTCCAGGAATCGAAGTCAAAGTACACTATGCAGGCTTATCGAAGATGGATGTATCAAAAGCTTTGGGAGAAAGTATCGTCATAAATTCGAATGTCAATTGTTTTGGATTTGACTTTTCCGGGGTCACAAGAAGGTAA
Protein
MSEHSKVATGFIIKTKVVNQCINEVDSDHKDMELNADDIYHLLKLRDYSYSGDFRSVHNGNSILNSFSIIWNDNWVTFIDGLLQVNALRQRHDGVSQPAEIRKLCIDVTEHRKHITKVDNKEIIPAKVLEKHETTKAGGVLLQGLLFQQLPPVILKGICLKSSKFIPRFNAKLDDVSAIHVFLQIVMENIDKDELNVLGILNDSGLVHPALAKITNEFNGISIHYKELSRKILITKKSEHLADADLVVVRGMTTDSCGAFGGYYYLPLKQEATKEDCELKLECSVTGNLNSLKWVQNSQSSTPGIEVKVHYAGLSKMDVSKALGESIVINSNVNCFGFDFSGVTRR
Summary
Uniprot
EMBL
ODYU01003708
SOQ42815.1
JTDY01003591
KOB69292.1
NWSH01000156
PCG78967.1
+ More
PCG78966.1 JTDY01000102 KOB78858.1 KQ459602 KPI93194.1 AGBW02011888 OWR45966.1 RSAL01000013 RVE53445.1 KQ461178 KPJ07769.1 BABH01033622 BABH01033623 BABH01033624 CH964272 EDW84866.1 GGMR01010533 MBY23152.1 JTDY01003650 KOB69211.1
PCG78966.1 JTDY01000102 KOB78858.1 KQ459602 KPI93194.1 AGBW02011888 OWR45966.1 RSAL01000013 RVE53445.1 KQ461178 KPJ07769.1 BABH01033622 BABH01033623 BABH01033624 CH964272 EDW84866.1 GGMR01010533 MBY23152.1 JTDY01003650 KOB69211.1
Proteomes
PRIDE
Pfam
Interpro
IPR020807
PKS_dehydratase
+ More
IPR020843 PKS_ER
IPR020801 PKS_acyl_transferase
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR001227 Ac_transferase_dom_sf
IPR032821 KAsynt_C_assoc
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR013968 PKS_KR
IPR013149 ADH_C
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
IPR020843 PKS_ER
IPR020801 PKS_acyl_transferase
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR001227 Ac_transferase_dom_sf
IPR032821 KAsynt_C_assoc
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR013968 PKS_KR
IPR013149 ADH_C
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
SUPFAM
CDD
ProteinModelPortal
PDB
6FN6
E-value=3.66433e-09,
Score=146
Ontologies
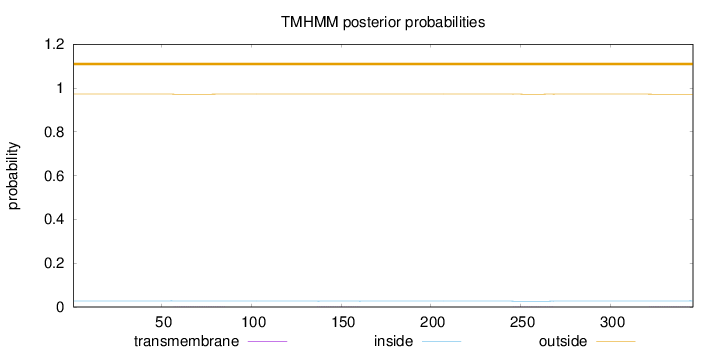
Topology
Length:
346
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06013
Exp number, first 60 AAs:
0.00091
Total prob of N-in:
0.02778
outside
1 - 346
Population Genetic Test Statistics
Pi
270.89555
Theta
198.948596
Tajima's D
1.227117
CLR
0
CSRT
0.723413829308535
Interpretation
Uncertain
