Gene
KWMTBOMO09837 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013131
Annotation
uncharacterized_protein_LOC106117886_precursor_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 0.995
Sequence
CDS
ATGTGGAAGTTGACGGTATTAGGAGTGTTTTTGACTGTTACCTATGTTTACAGTCACACTTATCACTTAGGATCATGTCCCATTGTTGAGCCTATGAGCGGATTTGAGATGAACAGGTTGCTAGGTGTGTGGTACGTTATACAGAAGACCTCAACAGCATCCCACTGCATCACGTACAACTTTAGCAGGACCGACGAGCCTGGACAATATCAGCTGGAACAGGACTCTCAACATTTCATACTCGGATTGACGCCCTTGAAACACGACTACAAATATACTGGTGTCCTGACAGTACCTGACCCCGCTGTTCCTGCAAGGATGAAGGTTCGATTCCCTCTGAGCGTTGCCGGCTCAGCAAGCTACACAGTTTTGGCAACGGACTACACAACATACGCCGCGGTCTTCACTTGTCAGAAGTTAGCGTTCGCTCATAGACGATCTGCCACTATCCTCTCGAGAACGAAAGAACTCGACAAAATGTTTGTTGATAAAATGCGTTTAAAGCTTTCTTCGTATGGAGTGGACCCTTATGATCTTTCGATCATATCACAGAACGATTGTCCAGGTCTACCCGATGGCGCATCTGAAGGCGTGAACATAGATATAAACCCCGAGACTTTCTCGTCTCACAACATCGGCGAAGCTGTCAGGAAAACTGGCGGAGCTATTGCTGACGGCGTGGAATACGTTGTGGAGGCTGGAAAGAAGGTGTACCACAAAGTGGCTAGCAGCAAGGAGGACCTTACCGAACCACCTTCGAACCCATACGCAGTCAGACCTATGGACAGCGATGCCGAGTGGCTACCCTAA
Protein
MWKLTVLGVFLTVTYVYSHTYHLGSCPIVEPMSGFEMNRLLGVWYVIQKTSTASHCITYNFSRTDEPGQYQLEQDSQHFILGLTPLKHDYKYTGVLTVPDPAVPARMKVRFPLSVAGSASYTVLATDYTTYAAVFTCQKLAFAHRRSATILSRTKELDKMFVDKMRLKLSSYGVDPYDLSIISQNDCPGLPDGASEGVNIDINPETFSSHNIGEAVRKTGGAIADGVEYVVEAGKKVYHKVASSKEDLTEPPSNPYAVRPMDSDAEWLP
Summary
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Uniprot
S4P564
I4DJX8
A0A194PIP4
I4DMP6
A0A3S2PKF8
A0A1E1WPJ5
+ More
A0A212F279 A0A2H1VNR8 H9JUB8 A0A0N1PEX1 A0A2A4K9P5 A0A0N1IQN8 A0A0K8TRK0 A0A195F283 A0A0J7L7B9 F4WI77 D2A5L9 K7IS12 A0A195BEJ9 A0A195CNX2 A0A158NZS0 E2AJQ0 A0A151IXU0 A0A154PND1 D2XP15 A0A151WV75 A0A088A4R2 A0A026WJ37 D2XP16 A0A3L8DY12 V9IAI3 A0A088A4V4 A0A2A3E9C1 A0A1L8DPU5 A0A0M8ZRN8 N6URW7 E2BR45 A0A310SSP7 A0A1B6CYH2 A0A1B6D487 Q16IR4 A0A1B6GWE0 A0A023END6 A0A1Y1KDK4 A0A1B0EX80 A0A0L7R393 A0A2M4BVK4 T1E9U7 A0A2M4CJJ2 A0A182R8N5 A0A224XMJ3 A0A182FPP4 A0A2M4AB47 A0A2M4ABX3 A0A2M3ZL17 A0A182WF74 Q16IR3 A0A182XC85 A0A336L2W5 A0A182VGW4 A0A171AZX3 A7UUU6 A0A182HNI7 A0A182YPD8 R4G3B5 A0A2M4CL24 A0A1B0CDV4 A0A336MRS7 A0A1Q3FIT1 A0A182MY02 A0A1L8DQ05 A0A182LPG1 U5EYU0 A0A2P8YQH8 B0WI31 A0A067RK92 A0A146KUD1 A0A0A9XKF9 A0A2M4BX64 A0A1J1HKQ5 A0A1Q3FIQ5 A0A2M4CKV3 A0A2M4AD06 A0A2M4ACJ4 A0A1Q1NP50 A0A182IS45 A0A1Q3FID5 A0A1Q3FIF2 A0A2M4BYB7 A0A2H8TSE2 A0A1Q3FIH7 A0A1Q3FIQ6 A0A182QI57 A0A2J7Q716 A0A2S2R9Z1 A0A2S2NNE8 J9JS80 A0A084WBC2 A0A1Y1KDY6 A0A1B6L058
A0A212F279 A0A2H1VNR8 H9JUB8 A0A0N1PEX1 A0A2A4K9P5 A0A0N1IQN8 A0A0K8TRK0 A0A195F283 A0A0J7L7B9 F4WI77 D2A5L9 K7IS12 A0A195BEJ9 A0A195CNX2 A0A158NZS0 E2AJQ0 A0A151IXU0 A0A154PND1 D2XP15 A0A151WV75 A0A088A4R2 A0A026WJ37 D2XP16 A0A3L8DY12 V9IAI3 A0A088A4V4 A0A2A3E9C1 A0A1L8DPU5 A0A0M8ZRN8 N6URW7 E2BR45 A0A310SSP7 A0A1B6CYH2 A0A1B6D487 Q16IR4 A0A1B6GWE0 A0A023END6 A0A1Y1KDK4 A0A1B0EX80 A0A0L7R393 A0A2M4BVK4 T1E9U7 A0A2M4CJJ2 A0A182R8N5 A0A224XMJ3 A0A182FPP4 A0A2M4AB47 A0A2M4ABX3 A0A2M3ZL17 A0A182WF74 Q16IR3 A0A182XC85 A0A336L2W5 A0A182VGW4 A0A171AZX3 A7UUU6 A0A182HNI7 A0A182YPD8 R4G3B5 A0A2M4CL24 A0A1B0CDV4 A0A336MRS7 A0A1Q3FIT1 A0A182MY02 A0A1L8DQ05 A0A182LPG1 U5EYU0 A0A2P8YQH8 B0WI31 A0A067RK92 A0A146KUD1 A0A0A9XKF9 A0A2M4BX64 A0A1J1HKQ5 A0A1Q3FIQ5 A0A2M4CKV3 A0A2M4AD06 A0A2M4ACJ4 A0A1Q1NP50 A0A182IS45 A0A1Q3FID5 A0A1Q3FIF2 A0A2M4BYB7 A0A2H8TSE2 A0A1Q3FIH7 A0A1Q3FIQ6 A0A182QI57 A0A2J7Q716 A0A2S2R9Z1 A0A2S2NNE8 J9JS80 A0A084WBC2 A0A1Y1KDY6 A0A1B6L058
Pubmed
EMBL
GAIX01007341
JAA85219.1
AK401596
BAM18218.1
KQ459602
KPI93192.1
+ More
AK402564 BAM19186.1 RSAL01000008 RVE53973.1 GDQN01002303 GDQN01001538 JAT88751.1 JAT89516.1 AGBW02010783 OWR47821.1 ODYU01003558 SOQ42475.1 BABH01038528 BABH01038529 BABH01038530 KQ461178 KPJ07771.1 NWSH01000035 PCG80450.1 LADJ01044234 KPJ21413.1 GDAI01000609 JAI16994.1 KQ981856 KYN34685.1 LBMM01000393 KMQ98491.1 GL888175 EGI66008.1 KQ971345 EFA05391.1 KQ976511 KYM82590.1 KQ977580 KYN01799.1 ADTU01004932 GL440062 EFN66334.1 KQ980791 KYN13116.1 KQ434999 KZC13376.1 GU228487 ADA82597.1 KQ982717 KYQ51728.1 KK107182 EZA56013.1 GU228488 ADA82598.1 QOIP01000003 RLU24708.1 JR038022 JR038023 AEY58084.1 AEY58085.1 KZ288315 PBC28325.1 GFDF01005602 JAV08482.1 KQ435885 KOX69577.1 APGK01019170 KB740092 KB632310 ENN81477.1 ERL92100.1 GL449898 EFN81851.1 KQ760801 OAD58956.1 GEDC01018923 JAS18375.1 GEDC01016787 JAS20511.1 CH478058 EAT34158.1 GECZ01009113 GECZ01003104 JAS60656.1 JAS66665.1 GAPW01003178 GAPW01003177 JAC10420.1 GEZM01087883 JAV58421.1 AJVK01000925 KQ414663 KOC65251.1 GGFJ01007975 MBW57116.1 GAMD01000992 JAB00599.1 GGFL01001316 MBW65494.1 GFTR01004122 JAW12304.1 GGFK01004658 MBW37979.1 GGFK01004787 MBW38108.1 GGFM01008357 MBW29108.1 EAT34157.1 UFQS01001769 UFQT01001769 SSX12286.1 SSX31737.1 GEMB01000447 JAS02678.1 AAAB01008966 EDO63529.1 APCN01001936 ACPB03000923 GAHY01002345 JAA75165.1 GGFL01001440 MBW65618.1 AJWK01008281 UFQT01001784 SSX31799.1 GFDL01007643 JAV27402.1 GFDF01005603 JAV08481.1 GANO01000126 JAB59745.1 PYGN01000431 PSN46512.1 DS231941 EDS28113.1 KK852575 KDR21026.1 GDHC01018535 JAQ00094.1 GBHO01022397 JAG21207.1 GGFJ01008482 MBW57623.1 CVRI01000006 CRK88114.1 GFDL01007629 JAV27416.1 GGFL01001802 MBW65980.1 GGFK01005346 MBW38667.1 GGFK01005182 MBW38503.1 KX752802 AQM58296.1 GFDL01007694 JAV27351.1 GFDL01007698 JAV27347.1 GGFJ01008936 MBW58077.1 GFXV01005331 MBW17136.1 GFDL01007681 JAV27364.1 GFDL01007673 JAV27372.1 AXCN02001648 NEVH01017450 PNF24358.1 GGMS01017287 MBY86490.1 GGMR01005843 MBY18462.1 ABLF02036422 ATLV01022343 KE525331 KFB47516.1 GEZM01087884 JAV58420.1 GEBQ01022895 JAT17082.1
AK402564 BAM19186.1 RSAL01000008 RVE53973.1 GDQN01002303 GDQN01001538 JAT88751.1 JAT89516.1 AGBW02010783 OWR47821.1 ODYU01003558 SOQ42475.1 BABH01038528 BABH01038529 BABH01038530 KQ461178 KPJ07771.1 NWSH01000035 PCG80450.1 LADJ01044234 KPJ21413.1 GDAI01000609 JAI16994.1 KQ981856 KYN34685.1 LBMM01000393 KMQ98491.1 GL888175 EGI66008.1 KQ971345 EFA05391.1 KQ976511 KYM82590.1 KQ977580 KYN01799.1 ADTU01004932 GL440062 EFN66334.1 KQ980791 KYN13116.1 KQ434999 KZC13376.1 GU228487 ADA82597.1 KQ982717 KYQ51728.1 KK107182 EZA56013.1 GU228488 ADA82598.1 QOIP01000003 RLU24708.1 JR038022 JR038023 AEY58084.1 AEY58085.1 KZ288315 PBC28325.1 GFDF01005602 JAV08482.1 KQ435885 KOX69577.1 APGK01019170 KB740092 KB632310 ENN81477.1 ERL92100.1 GL449898 EFN81851.1 KQ760801 OAD58956.1 GEDC01018923 JAS18375.1 GEDC01016787 JAS20511.1 CH478058 EAT34158.1 GECZ01009113 GECZ01003104 JAS60656.1 JAS66665.1 GAPW01003178 GAPW01003177 JAC10420.1 GEZM01087883 JAV58421.1 AJVK01000925 KQ414663 KOC65251.1 GGFJ01007975 MBW57116.1 GAMD01000992 JAB00599.1 GGFL01001316 MBW65494.1 GFTR01004122 JAW12304.1 GGFK01004658 MBW37979.1 GGFK01004787 MBW38108.1 GGFM01008357 MBW29108.1 EAT34157.1 UFQS01001769 UFQT01001769 SSX12286.1 SSX31737.1 GEMB01000447 JAS02678.1 AAAB01008966 EDO63529.1 APCN01001936 ACPB03000923 GAHY01002345 JAA75165.1 GGFL01001440 MBW65618.1 AJWK01008281 UFQT01001784 SSX31799.1 GFDL01007643 JAV27402.1 GFDF01005603 JAV08481.1 GANO01000126 JAB59745.1 PYGN01000431 PSN46512.1 DS231941 EDS28113.1 KK852575 KDR21026.1 GDHC01018535 JAQ00094.1 GBHO01022397 JAG21207.1 GGFJ01008482 MBW57623.1 CVRI01000006 CRK88114.1 GFDL01007629 JAV27416.1 GGFL01001802 MBW65980.1 GGFK01005346 MBW38667.1 GGFK01005182 MBW38503.1 KX752802 AQM58296.1 GFDL01007694 JAV27351.1 GFDL01007698 JAV27347.1 GGFJ01008936 MBW58077.1 GFXV01005331 MBW17136.1 GFDL01007681 JAV27364.1 GFDL01007673 JAV27372.1 AXCN02001648 NEVH01017450 PNF24358.1 GGMS01017287 MBY86490.1 GGMR01005843 MBY18462.1 ABLF02036422 ATLV01022343 KE525331 KFB47516.1 GEZM01087884 JAV58420.1 GEBQ01022895 JAT17082.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000005204
UP000053240
UP000218220
+ More
UP000078541 UP000036403 UP000007755 UP000007266 UP000002358 UP000078540 UP000078542 UP000005205 UP000000311 UP000078492 UP000076502 UP000075809 UP000005203 UP000053097 UP000279307 UP000242457 UP000053105 UP000019118 UP000030742 UP000008237 UP000008820 UP000092462 UP000053825 UP000075900 UP000069272 UP000075920 UP000076407 UP000075903 UP000007062 UP000075840 UP000076408 UP000015103 UP000092461 UP000075884 UP000075882 UP000245037 UP000002320 UP000027135 UP000183832 UP000075880 UP000075886 UP000235965 UP000007819 UP000030765
UP000078541 UP000036403 UP000007755 UP000007266 UP000002358 UP000078540 UP000078542 UP000005205 UP000000311 UP000078492 UP000076502 UP000075809 UP000005203 UP000053097 UP000279307 UP000242457 UP000053105 UP000019118 UP000030742 UP000008237 UP000008820 UP000092462 UP000053825 UP000075900 UP000069272 UP000075920 UP000076407 UP000075903 UP000007062 UP000075840 UP000076408 UP000015103 UP000092461 UP000075884 UP000075882 UP000245037 UP000002320 UP000027135 UP000183832 UP000075880 UP000075886 UP000235965 UP000007819 UP000030765
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
S4P564
I4DJX8
A0A194PIP4
I4DMP6
A0A3S2PKF8
A0A1E1WPJ5
+ More
A0A212F279 A0A2H1VNR8 H9JUB8 A0A0N1PEX1 A0A2A4K9P5 A0A0N1IQN8 A0A0K8TRK0 A0A195F283 A0A0J7L7B9 F4WI77 D2A5L9 K7IS12 A0A195BEJ9 A0A195CNX2 A0A158NZS0 E2AJQ0 A0A151IXU0 A0A154PND1 D2XP15 A0A151WV75 A0A088A4R2 A0A026WJ37 D2XP16 A0A3L8DY12 V9IAI3 A0A088A4V4 A0A2A3E9C1 A0A1L8DPU5 A0A0M8ZRN8 N6URW7 E2BR45 A0A310SSP7 A0A1B6CYH2 A0A1B6D487 Q16IR4 A0A1B6GWE0 A0A023END6 A0A1Y1KDK4 A0A1B0EX80 A0A0L7R393 A0A2M4BVK4 T1E9U7 A0A2M4CJJ2 A0A182R8N5 A0A224XMJ3 A0A182FPP4 A0A2M4AB47 A0A2M4ABX3 A0A2M3ZL17 A0A182WF74 Q16IR3 A0A182XC85 A0A336L2W5 A0A182VGW4 A0A171AZX3 A7UUU6 A0A182HNI7 A0A182YPD8 R4G3B5 A0A2M4CL24 A0A1B0CDV4 A0A336MRS7 A0A1Q3FIT1 A0A182MY02 A0A1L8DQ05 A0A182LPG1 U5EYU0 A0A2P8YQH8 B0WI31 A0A067RK92 A0A146KUD1 A0A0A9XKF9 A0A2M4BX64 A0A1J1HKQ5 A0A1Q3FIQ5 A0A2M4CKV3 A0A2M4AD06 A0A2M4ACJ4 A0A1Q1NP50 A0A182IS45 A0A1Q3FID5 A0A1Q3FIF2 A0A2M4BYB7 A0A2H8TSE2 A0A1Q3FIH7 A0A1Q3FIQ6 A0A182QI57 A0A2J7Q716 A0A2S2R9Z1 A0A2S2NNE8 J9JS80 A0A084WBC2 A0A1Y1KDY6 A0A1B6L058
A0A212F279 A0A2H1VNR8 H9JUB8 A0A0N1PEX1 A0A2A4K9P5 A0A0N1IQN8 A0A0K8TRK0 A0A195F283 A0A0J7L7B9 F4WI77 D2A5L9 K7IS12 A0A195BEJ9 A0A195CNX2 A0A158NZS0 E2AJQ0 A0A151IXU0 A0A154PND1 D2XP15 A0A151WV75 A0A088A4R2 A0A026WJ37 D2XP16 A0A3L8DY12 V9IAI3 A0A088A4V4 A0A2A3E9C1 A0A1L8DPU5 A0A0M8ZRN8 N6URW7 E2BR45 A0A310SSP7 A0A1B6CYH2 A0A1B6D487 Q16IR4 A0A1B6GWE0 A0A023END6 A0A1Y1KDK4 A0A1B0EX80 A0A0L7R393 A0A2M4BVK4 T1E9U7 A0A2M4CJJ2 A0A182R8N5 A0A224XMJ3 A0A182FPP4 A0A2M4AB47 A0A2M4ABX3 A0A2M3ZL17 A0A182WF74 Q16IR3 A0A182XC85 A0A336L2W5 A0A182VGW4 A0A171AZX3 A7UUU6 A0A182HNI7 A0A182YPD8 R4G3B5 A0A2M4CL24 A0A1B0CDV4 A0A336MRS7 A0A1Q3FIT1 A0A182MY02 A0A1L8DQ05 A0A182LPG1 U5EYU0 A0A2P8YQH8 B0WI31 A0A067RK92 A0A146KUD1 A0A0A9XKF9 A0A2M4BX64 A0A1J1HKQ5 A0A1Q3FIQ5 A0A2M4CKV3 A0A2M4AD06 A0A2M4ACJ4 A0A1Q1NP50 A0A182IS45 A0A1Q3FID5 A0A1Q3FIF2 A0A2M4BYB7 A0A2H8TSE2 A0A1Q3FIH7 A0A1Q3FIQ6 A0A182QI57 A0A2J7Q716 A0A2S2R9Z1 A0A2S2NNE8 J9JS80 A0A084WBC2 A0A1Y1KDY6 A0A1B6L058
PDB
2HZR
E-value=8.72212e-13,
Score=176
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.964526
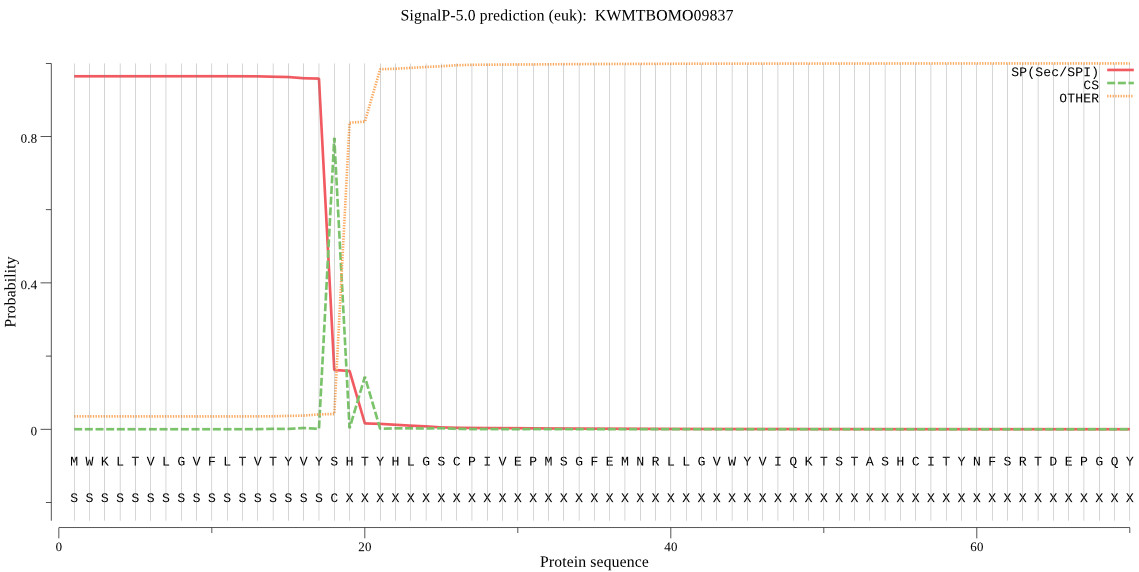
Length:
269
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.79611
Exp number, first 60 AAs:
4.51986
Total prob of N-in:
0.21311
outside
1 - 269
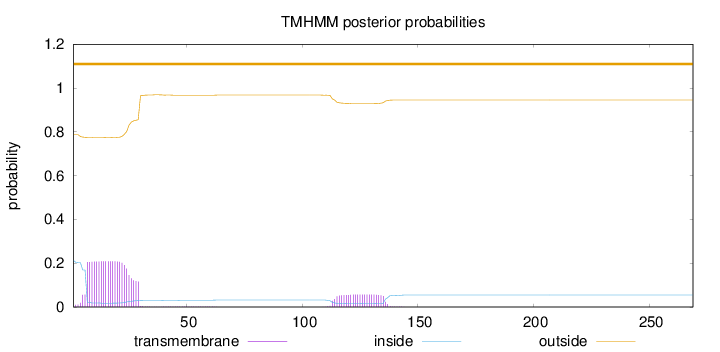
Population Genetic Test Statistics
Pi
129.467964
Theta
135.065061
Tajima's D
0.207119
CLR
0.23384
CSRT
0.424278786060697
Interpretation
Uncertain
