Gene
KWMTBOMO09836 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013130
Annotation
PREDICTED:_glucosylceramidase-like_isoform_X2_[Bombyx_mori]
Full name
Glucosylceramidase
Location in the cell
Cytoplasmic Reliability : 1.238 Extracellular Reliability : 1.211
Sequence
CDS
ATGGACTTGAACTATAACTACACCGGTTGGATAGATTGGAATTTGTGTCTCGATACTGAAGGTGGTCCAAATTGGGTCCAAAATTATGTCGACTCGCCGATTGTTGTCGACGCGGAGGCAGGTGTCTTCTACAAGCAGCCGATATTCTATGCTATGGGTCATTTTTCTAAGTTCATCCCTAGAGGCTCGAGGAGGATCAAGTCTACGGAGAAATATAACTGTTCAAACGCTTTGAAACATGTAGCCTTTTTGACACCAGAAAATACTATCGTCGTGGTTCTTCTTAACGAAGATAACAAAGACAGAGTTATTCGTTTGAAGTTCGGTGACAATCAAGCGAAGGTGCTTGTCGAGGGAAAGTCTATAATCACCGTCGAGTTCAACAACGAAGTTCAAGACGATTCTTGCGACTGTGATTGA
Protein
MDLNYNYTGWIDWNLCLDTEGGPNWVQNYVDSPIVVDAEAGVFYKQPIFYAMGHFSKFIPRGSRRIKSTEKYNCSNALKHVAFLTPENTIVVVLLNEDNKDRVIRLKFGDNQAKVLVEGKSIITVEFNNEVQDDSCDCD
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
H9JUB7
H9JUB6
A0A0N1INL2
A0A2A4K975
A0A212EPK9
A0A437ARV3
+ More
A0A0N1IF15 A0A2A4K8P8 A0A2H1WWW7 A0A437BUD8 A0A1W1EGM7 A0A212F349 A0A212F293 A0A2H1VST3 A0A2H1VUS0 A0A3M0J565 A0A131Z3G7 A0A212F254 L7M0B5 A0A2H1VG50 H3G7I4 A0A452ICC8 A0A023GIV4 L7M0C6 A0A131YKA6 A0A3L8RV84 A0A1V4KDC5 A0A224YYV0 A0A224YXV5 G4ZEY4 A0A329SI21 A0A093BHB6 A0A218UMU9 A0A452ICI5 A0A437B0B5 A0A2I0LM54 A0A1D5NWE6 A0A3Q2U6X4 W2QN93 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 A0A131YJ49 A0A2G9U5T9 W2ZX75 A0A093GAN8 A0A225WY05 U3JJS0 A0A3L8RW53 A0A091I5V7 A0A3R7MN95 A0A293MAW8 A0A1U7SIA2 A0A329T1H5 A0A452ICH6 A0A081ANK4 A0A452ID18 V9FJP4 W2XFZ2 W2QJE5 W2ZQ36 W2HAK1 U3JJS6 A0A421G9R3 A0A3Q0HD48 A0A3Q0DMP0 A0A091FYT6 A0A3M0IZB8 A0A1U7SVT9 J9L5B0 A0A2P4WZX2 A0A1E1X9A0 A0A3Q0HA13 A0A1U7T338 A0A3Q0HD38 A0A293N7G8 W2HHH0 A0A151N939 A0A3R7H2D6 W2JP47 A0A0C9QXG4 A0A0Q3QSH3 A0A218UMY8 A0A099ZCW2
A0A0N1IF15 A0A2A4K8P8 A0A2H1WWW7 A0A437BUD8 A0A1W1EGM7 A0A212F349 A0A212F293 A0A2H1VST3 A0A2H1VUS0 A0A3M0J565 A0A131Z3G7 A0A212F254 L7M0B5 A0A2H1VG50 H3G7I4 A0A452ICC8 A0A023GIV4 L7M0C6 A0A131YKA6 A0A3L8RV84 A0A1V4KDC5 A0A224YYV0 A0A224YXV5 G4ZEY4 A0A329SI21 A0A093BHB6 A0A218UMU9 A0A452ICI5 A0A437B0B5 A0A2I0LM54 A0A1D5NWE6 A0A3Q2U6X4 W2QN93 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 A0A131YJ49 A0A2G9U5T9 W2ZX75 A0A093GAN8 A0A225WY05 U3JJS0 A0A3L8RW53 A0A091I5V7 A0A3R7MN95 A0A293MAW8 A0A1U7SIA2 A0A329T1H5 A0A452ICH6 A0A081ANK4 A0A452ID18 V9FJP4 W2XFZ2 W2QJE5 W2ZQ36 W2HAK1 U3JJS6 A0A421G9R3 A0A3Q0HD48 A0A3Q0DMP0 A0A091FYT6 A0A3M0IZB8 A0A1U7SVT9 J9L5B0 A0A2P4WZX2 A0A1E1X9A0 A0A3Q0HA13 A0A1U7T338 A0A3Q0HD38 A0A293N7G8 W2HHH0 A0A151N939 A0A3R7H2D6 W2JP47 A0A0C9QXG4 A0A0Q3QSH3 A0A218UMY8 A0A099ZCW2
EC Number
3.2.1.45
Pubmed
EMBL
BABH01038531
BABH01038532
BABH01038533
BABH01038534
BABH01038539
BABH01038540
+ More
BABH01038541 KQ461178 KPJ07772.1 NWSH01000035 PCG80448.1 AGBW02013448 OWR43423.1 RSAL01003486 RVE40276.1 KPJ07773.1 PCG80449.1 ODYU01011672 SOQ57565.1 RSAL01000008 RVE53975.1 LT635665 SGZ49395.1 AGBW02010629 OWR48154.1 AGBW02010783 OWR47849.1 ODYU01004232 SOQ43893.1 ODYU01004571 SOQ44585.1 QRBI01000202 RMB93899.1 GEDV01003886 JAP84671.1 OWR47828.1 GACK01007727 JAA57307.1 ODYU01002190 SOQ39362.1 DS566059 GBBM01001526 JAC33892.1 GACK01007712 JAA57322.1 GEDV01009569 JAP78988.1 QUSF01000198 RLV87492.1 LSYS01003627 OPJ82428.1 GFPF01008008 MAA19154.1 GFPF01008007 MAA19153.1 JH159154 EGZ17896.1 MJFZ01000176 RAW35212.1 KN125716 KFU83958.1 MUZQ01000219 OWK55107.1 RSAL01000232 RVE43819.1 AKCR02000200 PKK18507.1 AADN05000466 KI669572 ETN14662.1 ANIZ01000561 ETI54139.1 ANIX01000668 ETP23960.1 ANJA01000623 ETO82874.1 KI678010 KI691249 ETM00502.1 ETM53704.1 LNFP01001456 LNFO01001593 KUF86242.1 KUF90004.1 GEDV01009570 JAP78987.1 KZ349327 PIO65072.1 ANIY01000684 ETP51967.1 KL215229 KFV63809.1 NBNE01000176 OWZ21918.1 AGTO01022043 RLV87493.1 KL218155 KFP02780.1 MAYM02001651 RLN14102.1 GFWV01011406 MAA36135.1 MJFZ01000012 RAW42725.1 ANJA01000988 ETO80465.1 ANIZ01000889 ETI51715.1 ANIX01001073 ETP21412.1 KI669577 ETN12365.1 ANIY01001102 ETP49403.1 KI685308 KI671854 KI678592 KI691839 ETK91605.1 ETL45015.1 ETL98169.1 ETM51333.1 MBAD02000201 RLN70671.1 KL447511 KFO74306.1 RMB93898.1 ABLF02008893 NCKW01020142 POM58843.1 GFAC01003537 JAT95651.1 GFWV01022833 MAA47560.1 KI684753 ETK94015.1 AKHW03003703 KYO33342.1 MBAD02001844 RLN51535.1 KI671251 ETL47408.1 GBYB01005352 JAG75119.1 LMAW01002883 KQK76089.1 OWK55104.1 KL892965 KGL80274.1
BABH01038541 KQ461178 KPJ07772.1 NWSH01000035 PCG80448.1 AGBW02013448 OWR43423.1 RSAL01003486 RVE40276.1 KPJ07773.1 PCG80449.1 ODYU01011672 SOQ57565.1 RSAL01000008 RVE53975.1 LT635665 SGZ49395.1 AGBW02010629 OWR48154.1 AGBW02010783 OWR47849.1 ODYU01004232 SOQ43893.1 ODYU01004571 SOQ44585.1 QRBI01000202 RMB93899.1 GEDV01003886 JAP84671.1 OWR47828.1 GACK01007727 JAA57307.1 ODYU01002190 SOQ39362.1 DS566059 GBBM01001526 JAC33892.1 GACK01007712 JAA57322.1 GEDV01009569 JAP78988.1 QUSF01000198 RLV87492.1 LSYS01003627 OPJ82428.1 GFPF01008008 MAA19154.1 GFPF01008007 MAA19153.1 JH159154 EGZ17896.1 MJFZ01000176 RAW35212.1 KN125716 KFU83958.1 MUZQ01000219 OWK55107.1 RSAL01000232 RVE43819.1 AKCR02000200 PKK18507.1 AADN05000466 KI669572 ETN14662.1 ANIZ01000561 ETI54139.1 ANIX01000668 ETP23960.1 ANJA01000623 ETO82874.1 KI678010 KI691249 ETM00502.1 ETM53704.1 LNFP01001456 LNFO01001593 KUF86242.1 KUF90004.1 GEDV01009570 JAP78987.1 KZ349327 PIO65072.1 ANIY01000684 ETP51967.1 KL215229 KFV63809.1 NBNE01000176 OWZ21918.1 AGTO01022043 RLV87493.1 KL218155 KFP02780.1 MAYM02001651 RLN14102.1 GFWV01011406 MAA36135.1 MJFZ01000012 RAW42725.1 ANJA01000988 ETO80465.1 ANIZ01000889 ETI51715.1 ANIX01001073 ETP21412.1 KI669577 ETN12365.1 ANIY01001102 ETP49403.1 KI685308 KI671854 KI678592 KI691839 ETK91605.1 ETL45015.1 ETL98169.1 ETM51333.1 MBAD02000201 RLN70671.1 KL447511 KFO74306.1 RMB93898.1 ABLF02008893 NCKW01020142 POM58843.1 GFAC01003537 JAT95651.1 GFWV01022833 MAA47560.1 KI684753 ETK94015.1 AKHW03003703 KYO33342.1 MBAD02001844 RLN51535.1 KI671251 ETL47408.1 GBYB01005352 JAG75119.1 LMAW01002883 KQK76089.1 OWK55104.1 KL892965 KGL80274.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000283053
UP000269221
+ More
UP000005238 UP000291020 UP000276834 UP000190648 UP000002640 UP000251314 UP000197619 UP000053872 UP000000539 UP000018817 UP000018721 UP000018958 UP000028582 UP000054423 UP000054532 UP000052943 UP000018948 UP000053875 UP000198211 UP000016665 UP000054308 UP000285883 UP000189705 UP000053236 UP000053864 UP000284657 UP000189704 UP000053760 UP000007819 UP000050525 UP000051836 UP000053641
UP000005238 UP000291020 UP000276834 UP000190648 UP000002640 UP000251314 UP000197619 UP000053872 UP000000539 UP000018817 UP000018721 UP000018958 UP000028582 UP000054423 UP000054532 UP000052943 UP000018948 UP000053875 UP000198211 UP000016665 UP000054308 UP000285883 UP000189705 UP000053236 UP000053864 UP000284657 UP000189704 UP000053760 UP000007819 UP000050525 UP000051836 UP000053641
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JUB7
H9JUB6
A0A0N1INL2
A0A2A4K975
A0A212EPK9
A0A437ARV3
+ More
A0A0N1IF15 A0A2A4K8P8 A0A2H1WWW7 A0A437BUD8 A0A1W1EGM7 A0A212F349 A0A212F293 A0A2H1VST3 A0A2H1VUS0 A0A3M0J565 A0A131Z3G7 A0A212F254 L7M0B5 A0A2H1VG50 H3G7I4 A0A452ICC8 A0A023GIV4 L7M0C6 A0A131YKA6 A0A3L8RV84 A0A1V4KDC5 A0A224YYV0 A0A224YXV5 G4ZEY4 A0A329SI21 A0A093BHB6 A0A218UMU9 A0A452ICI5 A0A437B0B5 A0A2I0LM54 A0A1D5NWE6 A0A3Q2U6X4 W2QN93 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 A0A131YJ49 A0A2G9U5T9 W2ZX75 A0A093GAN8 A0A225WY05 U3JJS0 A0A3L8RW53 A0A091I5V7 A0A3R7MN95 A0A293MAW8 A0A1U7SIA2 A0A329T1H5 A0A452ICH6 A0A081ANK4 A0A452ID18 V9FJP4 W2XFZ2 W2QJE5 W2ZQ36 W2HAK1 U3JJS6 A0A421G9R3 A0A3Q0HD48 A0A3Q0DMP0 A0A091FYT6 A0A3M0IZB8 A0A1U7SVT9 J9L5B0 A0A2P4WZX2 A0A1E1X9A0 A0A3Q0HA13 A0A1U7T338 A0A3Q0HD38 A0A293N7G8 W2HHH0 A0A151N939 A0A3R7H2D6 W2JP47 A0A0C9QXG4 A0A0Q3QSH3 A0A218UMY8 A0A099ZCW2
A0A0N1IF15 A0A2A4K8P8 A0A2H1WWW7 A0A437BUD8 A0A1W1EGM7 A0A212F349 A0A212F293 A0A2H1VST3 A0A2H1VUS0 A0A3M0J565 A0A131Z3G7 A0A212F254 L7M0B5 A0A2H1VG50 H3G7I4 A0A452ICC8 A0A023GIV4 L7M0C6 A0A131YKA6 A0A3L8RV84 A0A1V4KDC5 A0A224YYV0 A0A224YXV5 G4ZEY4 A0A329SI21 A0A093BHB6 A0A218UMU9 A0A452ICI5 A0A437B0B5 A0A2I0LM54 A0A1D5NWE6 A0A3Q2U6X4 W2QN93 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 A0A131YJ49 A0A2G9U5T9 W2ZX75 A0A093GAN8 A0A225WY05 U3JJS0 A0A3L8RW53 A0A091I5V7 A0A3R7MN95 A0A293MAW8 A0A1U7SIA2 A0A329T1H5 A0A452ICH6 A0A081ANK4 A0A452ID18 V9FJP4 W2XFZ2 W2QJE5 W2ZQ36 W2HAK1 U3JJS6 A0A421G9R3 A0A3Q0HD48 A0A3Q0DMP0 A0A091FYT6 A0A3M0IZB8 A0A1U7SVT9 J9L5B0 A0A2P4WZX2 A0A1E1X9A0 A0A3Q0HA13 A0A1U7T338 A0A3Q0HD38 A0A293N7G8 W2HHH0 A0A151N939 A0A3R7H2D6 W2JP47 A0A0C9QXG4 A0A0Q3QSH3 A0A218UMY8 A0A099ZCW2
PDB
6Q6N
E-value=1.08978e-28,
Score=308
Ontologies
PATHWAY
GO
GO:0006665
GO:0004348
GO:0006680
GO:0016787
GO:0030259
GO:0071356
GO:0006914
GO:0023021
GO:1904457
GO:0005124
GO:0046527
GO:0032715
GO:0050295
GO:0005765
GO:0043243
GO:1903061
GO:0046512
GO:0007040
GO:0032463
GO:0008203
GO:0043407
GO:1901215
GO:0032006
GO:0046513
GO:1903052
GO:0035307
GO:0005975
GO:0008270
GO:0005488
GO:0008565
GO:0015031
GO:0009258
GO:0016155
GO:0003824
PANTHER
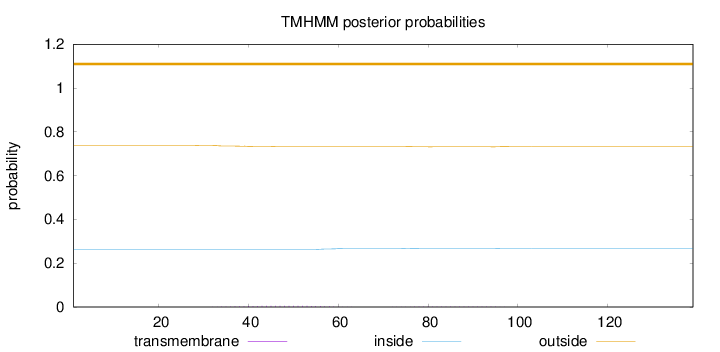
Topology
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15031
Exp number, first 60 AAs:
0.10318
Total prob of N-in:
0.26268
outside
1 - 139
Population Genetic Test Statistics
Pi
134.30763
Theta
124.631715
Tajima's D
0.056082
CLR
12.617959
CSRT
0.386530673466327
Interpretation
Uncertain
