Gene
KWMTBOMO09834
Pre Gene Modal
BGIBMGA013130
Annotation
glucosylceramidase_[Papilio_xuthus]
Full name
Glucosylceramidase
Location in the cell
PlasmaMembrane Reliability : 1.856
Sequence
CDS
ATGATTAGAACACCGATAGGAGGTAGTGATTTTTCAAGCCGCGCATACACTCTGAACGACTATCCTCTTGATGATAAGGAACTCGGAAACTTCAGTTTAAGTTACGAAGATTACAATTATAAGTTGCCAATGATAAAAGCTTGCCTAGCTGCAGCGAGCTCGCAGGTCTTCATGGTTGGAACAACGTGGTCACCTCCAGCGTGGATGAAGACTTCCGGATCATTAACGGGCGTCGGTTATCTTAAGGAAGAGTATTTTGAGGCGTATGCTAATTACCACAAGAAGTTCCTAGAAGAATACAAAAGGAACAACATTTCTTTTTGGGCTATAACGACCACTAATGAACCTTTGAATGGTGTTGTAGATCTACCGGACTTCAATTGTCTTGGTTGGACAATCGAGGGGATGGTGAGTGTAATGTCTTTTGTCGAGTTGGATTCTAAGCCCCAACGTCCCAGTTTTACAGTACAACGAATGCTCTGCTGCTAA
Protein
MIRTPIGGSDFSSRAYTLNDYPLDDKELGNFSLSYEDYNYKLPMIKACLAAASSQVFMVGTTWSPPAWMKTSGSLTGVGYLKEEYFEAYANYHKKFLEEYKRNNISFWAITTTNEPLNGVVDLPDFNCLGWTIEGMVSVMSFVELDSKPQRPSFTVQRMLCC
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
H9JUB7
I4DLH0
A0A212FNA8
A0A1W1EGM7
H9JUB6
A0A212F248
+ More
A0A194PPY1 A0A3S2NLJ3 A0A2H1VG50 A0A2H1VUS0 A0A194Q614 A0A0N1IF15 A0A0L7L523 A0A2H1WWW7 A0A2A4K8P8 A0A437B0B5 A0A2A4K975 A0A2H1VST3 A0A437BUD8 A0A2H1X380 J9K5F8 A0A3Q0JJ39 A0A0L7QMS7 E9HE79 A0A0P5V3Y6 A0A154P4F4 A0A0P5V259 A0A0P5V115 A0A0P5UAZ3 A0A0P5UEK4 A0A0P5MIQ6 T1L0X2 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0P5XZL7 D6WRN0 A0A0N8C3M9 A0A0P5VAG5 A0A0P5D4A0 A0A0P6HWY3 A0A0P5BCG5 A0A0P5BE18 A0A0N8BXP2 A0A0P5YAM8 A0A067QM95 A0A0P6CTA3 S4RMS8 A0A1B6LUZ4 A0A1D2MAR5 A0A0P5EBB8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 T1JW72 A0A2J7R6J1 A0A0P5E6V6 C3YQ03 A0A1B6FTD6 A0A0P5F794 A0A0N8EC25 A0A1B6IGC1 A0A1B6K1N7 A0A0P5UX07 A0A0P5YE76 A0A423SIH3 A0A0P4Y4V0 A0A0P6GJR3 A0A0N8DE38 A0A0P5XUP8 A0A0P5Y055 A0A0P5X813 A0A0P5YNS9 A0A139WEE1 A0A0P5V575 A0A0P5Y2L0 A0A0P5XL26 A0A162QRA1 A0A0N8A576 A0A0P5Z7Y2 A0A1Y3AW29 A0A0P5BMC7 A0A0P5L6K4 A0A0P5V0B5 A0A131Z9I7 A0A0P5WP50 A0A0P5F4K4 A0A0P5UYZ4 A0A0P5MXS5 A0A0L8GBA3 A0A0P6E3A7 A0A088APM4 A0A0P5E3J3 V9IBT9 A0A0P5GMD5 A0A226EKU6 A0A0P6HUV1 A0A0P6B2C4 A0A1D5NWE6 A0A224YYV0
A0A194PPY1 A0A3S2NLJ3 A0A2H1VG50 A0A2H1VUS0 A0A194Q614 A0A0N1IF15 A0A0L7L523 A0A2H1WWW7 A0A2A4K8P8 A0A437B0B5 A0A2A4K975 A0A2H1VST3 A0A437BUD8 A0A2H1X380 J9K5F8 A0A3Q0JJ39 A0A0L7QMS7 E9HE79 A0A0P5V3Y6 A0A154P4F4 A0A0P5V259 A0A0P5V115 A0A0P5UAZ3 A0A0P5UEK4 A0A0P5MIQ6 T1L0X2 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0P5XZL7 D6WRN0 A0A0N8C3M9 A0A0P5VAG5 A0A0P5D4A0 A0A0P6HWY3 A0A0P5BCG5 A0A0P5BE18 A0A0N8BXP2 A0A0P5YAM8 A0A067QM95 A0A0P6CTA3 S4RMS8 A0A1B6LUZ4 A0A1D2MAR5 A0A0P5EBB8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 T1JW72 A0A2J7R6J1 A0A0P5E6V6 C3YQ03 A0A1B6FTD6 A0A0P5F794 A0A0N8EC25 A0A1B6IGC1 A0A1B6K1N7 A0A0P5UX07 A0A0P5YE76 A0A423SIH3 A0A0P4Y4V0 A0A0P6GJR3 A0A0N8DE38 A0A0P5XUP8 A0A0P5Y055 A0A0P5X813 A0A0P5YNS9 A0A139WEE1 A0A0P5V575 A0A0P5Y2L0 A0A0P5XL26 A0A162QRA1 A0A0N8A576 A0A0P5Z7Y2 A0A1Y3AW29 A0A0P5BMC7 A0A0P5L6K4 A0A0P5V0B5 A0A131Z9I7 A0A0P5WP50 A0A0P5F4K4 A0A0P5UYZ4 A0A0P5MXS5 A0A0L8GBA3 A0A0P6E3A7 A0A088APM4 A0A0P5E3J3 V9IBT9 A0A0P5GMD5 A0A226EKU6 A0A0P6HUV1 A0A0P6B2C4 A0A1D5NWE6 A0A224YYV0
EC Number
3.2.1.45
Pubmed
EMBL
BABH01038531
BABH01038532
BABH01038533
BABH01038534
AK402138
BAM18760.1
+ More
AGBW02006772 OWR55189.1 LT635665 SGZ49395.1 BABH01038539 BABH01038540 BABH01038541 AGBW02010783 OWR47794.1 KQ459602 KPI93190.1 RSAL01000008 RVE53974.1 ODYU01002190 SOQ39362.1 ODYU01004571 SOQ44585.1 KQ459439 KPJ00977.1 KQ461178 KPJ07773.1 JTDY01002868 KOB70572.1 ODYU01011672 SOQ57565.1 NWSH01000035 PCG80449.1 RSAL01000232 RVE43819.1 PCG80448.1 ODYU01004232 SOQ43893.1 RVE53975.1 ODYU01013051 SOQ59702.1 ABLF02017156 ABLF02017157 ABLF02017158 ABLF02017160 ABLF02043031 ABLF02044170 KQ414868 KOC59952.1 GL732627 EFX69969.1 GDIP01105547 JAL98167.1 KQ434814 KZC06829.1 GDIP01105549 JAL98165.1 GDIP01105548 JAL98166.1 GDIP01115596 JAL88118.1 GDIP01114152 JAL89562.1 GDIQ01166874 JAK84851.1 CAEY01000873 OWR47828.1 GDIP01111638 JAL92076.1 GDIP01112839 JAL90875.1 GDIP01066853 JAM36862.1 KQ971354 EFA07058.2 GDIQ01118027 JAL33699.1 GDIP01102691 JAM01024.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01028607 JAN66130.1 GDIP01186374 JAJ37028.1 GDIP01190080 JAJ33322.1 GDIQ01134723 JAL17003.1 GDIP01074304 JAM29411.1 KK853153 KDR10552.1 GDIP01009481 JAM94234.1 GEBQ01024969 GEBQ01012479 JAT15008.1 JAT27498.1 LJIJ01002183 ODM90073.1 GDIP01148941 JAJ74461.1 NEVH01006756 PNF36451.1 GECU01013983 JAS93723.1 PNF36457.1 CAEY01000807 PNF36452.1 GDIP01147340 JAJ76062.1 GG666539 EEN57641.1 GECZ01016297 JAS53472.1 GDIQ01261732 JAJ89992.1 GDIQ01041468 JAN53269.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 GDIP01107443 JAL96271.1 GDIP01073106 JAM30609.1 QCYY01003347 ROT63991.1 GDIP01234780 JAI88621.1 GDIQ01034555 JAN60182.1 GDIP01042743 JAM60972.1 GDIP01067454 JAM36261.1 GDIP01065954 JAM37761.1 GDIP01077038 JAM26677.1 GDIP01056052 JAM47663.1 KYB26348.1 GDIP01104846 JAL98868.1 GDIP01077037 JAM26678.1 GDIP01070494 JAM33221.1 KZS19887.1 GDIP01181329 JAJ42073.1 GDIP01060297 JAM43418.1 MUJZ01055050 OTF72692.1 GDIP01182978 JAJ40424.1 GDIQ01199303 JAK52422.1 GDIP01107444 JAL96270.1 GEDV01001092 JAP87465.1 GDIP01083544 JAM20171.1 GDIQ01263085 JAJ88639.1 GDIP01107445 JAL96269.1 GDIQ01149727 JAL01999.1 KQ422986 KOF73820.1 GDIQ01068405 JAN26332.1 GDIP01147339 JAJ76063.1 JR037481 JR037482 AEY57789.1 AEY57790.1 GDIQ01261484 JAJ90240.1 LNIX01000003 OXA58323.1 GDIQ01038503 JAN56234.1 GDIP01023117 JAM80598.1 AADN05000466 GFPF01008008 MAA19154.1
AGBW02006772 OWR55189.1 LT635665 SGZ49395.1 BABH01038539 BABH01038540 BABH01038541 AGBW02010783 OWR47794.1 KQ459602 KPI93190.1 RSAL01000008 RVE53974.1 ODYU01002190 SOQ39362.1 ODYU01004571 SOQ44585.1 KQ459439 KPJ00977.1 KQ461178 KPJ07773.1 JTDY01002868 KOB70572.1 ODYU01011672 SOQ57565.1 NWSH01000035 PCG80449.1 RSAL01000232 RVE43819.1 PCG80448.1 ODYU01004232 SOQ43893.1 RVE53975.1 ODYU01013051 SOQ59702.1 ABLF02017156 ABLF02017157 ABLF02017158 ABLF02017160 ABLF02043031 ABLF02044170 KQ414868 KOC59952.1 GL732627 EFX69969.1 GDIP01105547 JAL98167.1 KQ434814 KZC06829.1 GDIP01105549 JAL98165.1 GDIP01105548 JAL98166.1 GDIP01115596 JAL88118.1 GDIP01114152 JAL89562.1 GDIQ01166874 JAK84851.1 CAEY01000873 OWR47828.1 GDIP01111638 JAL92076.1 GDIP01112839 JAL90875.1 GDIP01066853 JAM36862.1 KQ971354 EFA07058.2 GDIQ01118027 JAL33699.1 GDIP01102691 JAM01024.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01028607 JAN66130.1 GDIP01186374 JAJ37028.1 GDIP01190080 JAJ33322.1 GDIQ01134723 JAL17003.1 GDIP01074304 JAM29411.1 KK853153 KDR10552.1 GDIP01009481 JAM94234.1 GEBQ01024969 GEBQ01012479 JAT15008.1 JAT27498.1 LJIJ01002183 ODM90073.1 GDIP01148941 JAJ74461.1 NEVH01006756 PNF36451.1 GECU01013983 JAS93723.1 PNF36457.1 CAEY01000807 PNF36452.1 GDIP01147340 JAJ76062.1 GG666539 EEN57641.1 GECZ01016297 JAS53472.1 GDIQ01261732 JAJ89992.1 GDIQ01041468 JAN53269.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 GDIP01107443 JAL96271.1 GDIP01073106 JAM30609.1 QCYY01003347 ROT63991.1 GDIP01234780 JAI88621.1 GDIQ01034555 JAN60182.1 GDIP01042743 JAM60972.1 GDIP01067454 JAM36261.1 GDIP01065954 JAM37761.1 GDIP01077038 JAM26677.1 GDIP01056052 JAM47663.1 KYB26348.1 GDIP01104846 JAL98868.1 GDIP01077037 JAM26678.1 GDIP01070494 JAM33221.1 KZS19887.1 GDIP01181329 JAJ42073.1 GDIP01060297 JAM43418.1 MUJZ01055050 OTF72692.1 GDIP01182978 JAJ40424.1 GDIQ01199303 JAK52422.1 GDIP01107444 JAL96270.1 GEDV01001092 JAP87465.1 GDIP01083544 JAM20171.1 GDIQ01263085 JAJ88639.1 GDIP01107445 JAL96269.1 GDIQ01149727 JAL01999.1 KQ422986 KOF73820.1 GDIQ01068405 JAN26332.1 GDIP01147339 JAJ76063.1 JR037481 JR037482 AEY57789.1 AEY57790.1 GDIQ01261484 JAJ90240.1 LNIX01000003 OXA58323.1 GDIQ01038503 JAN56234.1 GDIP01023117 JAM80598.1 AADN05000466 GFPF01008008 MAA19154.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000218220 UP000007819 UP000079169 UP000053825 UP000000305 UP000076502 UP000015104 UP000007266 UP000076858 UP000027135 UP000245300 UP000094527 UP000235965 UP000001554 UP000283509 UP000053454 UP000005203 UP000198287 UP000000539
UP000218220 UP000007819 UP000079169 UP000053825 UP000000305 UP000076502 UP000015104 UP000007266 UP000076858 UP000027135 UP000245300 UP000094527 UP000235965 UP000001554 UP000283509 UP000053454 UP000005203 UP000198287 UP000000539
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JUB7
I4DLH0
A0A212FNA8
A0A1W1EGM7
H9JUB6
A0A212F248
+ More
A0A194PPY1 A0A3S2NLJ3 A0A2H1VG50 A0A2H1VUS0 A0A194Q614 A0A0N1IF15 A0A0L7L523 A0A2H1WWW7 A0A2A4K8P8 A0A437B0B5 A0A2A4K975 A0A2H1VST3 A0A437BUD8 A0A2H1X380 J9K5F8 A0A3Q0JJ39 A0A0L7QMS7 E9HE79 A0A0P5V3Y6 A0A154P4F4 A0A0P5V259 A0A0P5V115 A0A0P5UAZ3 A0A0P5UEK4 A0A0P5MIQ6 T1L0X2 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0P5XZL7 D6WRN0 A0A0N8C3M9 A0A0P5VAG5 A0A0P5D4A0 A0A0P6HWY3 A0A0P5BCG5 A0A0P5BE18 A0A0N8BXP2 A0A0P5YAM8 A0A067QM95 A0A0P6CTA3 S4RMS8 A0A1B6LUZ4 A0A1D2MAR5 A0A0P5EBB8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 T1JW72 A0A2J7R6J1 A0A0P5E6V6 C3YQ03 A0A1B6FTD6 A0A0P5F794 A0A0N8EC25 A0A1B6IGC1 A0A1B6K1N7 A0A0P5UX07 A0A0P5YE76 A0A423SIH3 A0A0P4Y4V0 A0A0P6GJR3 A0A0N8DE38 A0A0P5XUP8 A0A0P5Y055 A0A0P5X813 A0A0P5YNS9 A0A139WEE1 A0A0P5V575 A0A0P5Y2L0 A0A0P5XL26 A0A162QRA1 A0A0N8A576 A0A0P5Z7Y2 A0A1Y3AW29 A0A0P5BMC7 A0A0P5L6K4 A0A0P5V0B5 A0A131Z9I7 A0A0P5WP50 A0A0P5F4K4 A0A0P5UYZ4 A0A0P5MXS5 A0A0L8GBA3 A0A0P6E3A7 A0A088APM4 A0A0P5E3J3 V9IBT9 A0A0P5GMD5 A0A226EKU6 A0A0P6HUV1 A0A0P6B2C4 A0A1D5NWE6 A0A224YYV0
A0A194PPY1 A0A3S2NLJ3 A0A2H1VG50 A0A2H1VUS0 A0A194Q614 A0A0N1IF15 A0A0L7L523 A0A2H1WWW7 A0A2A4K8P8 A0A437B0B5 A0A2A4K975 A0A2H1VST3 A0A437BUD8 A0A2H1X380 J9K5F8 A0A3Q0JJ39 A0A0L7QMS7 E9HE79 A0A0P5V3Y6 A0A154P4F4 A0A0P5V259 A0A0P5V115 A0A0P5UAZ3 A0A0P5UEK4 A0A0P5MIQ6 T1L0X2 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0P5XZL7 D6WRN0 A0A0N8C3M9 A0A0P5VAG5 A0A0P5D4A0 A0A0P6HWY3 A0A0P5BCG5 A0A0P5BE18 A0A0N8BXP2 A0A0P5YAM8 A0A067QM95 A0A0P6CTA3 S4RMS8 A0A1B6LUZ4 A0A1D2MAR5 A0A0P5EBB8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 T1JW72 A0A2J7R6J1 A0A0P5E6V6 C3YQ03 A0A1B6FTD6 A0A0P5F794 A0A0N8EC25 A0A1B6IGC1 A0A1B6K1N7 A0A0P5UX07 A0A0P5YE76 A0A423SIH3 A0A0P4Y4V0 A0A0P6GJR3 A0A0N8DE38 A0A0P5XUP8 A0A0P5Y055 A0A0P5X813 A0A0P5YNS9 A0A139WEE1 A0A0P5V575 A0A0P5Y2L0 A0A0P5XL26 A0A162QRA1 A0A0N8A576 A0A0P5Z7Y2 A0A1Y3AW29 A0A0P5BMC7 A0A0P5L6K4 A0A0P5V0B5 A0A131Z9I7 A0A0P5WP50 A0A0P5F4K4 A0A0P5UYZ4 A0A0P5MXS5 A0A0L8GBA3 A0A0P6E3A7 A0A088APM4 A0A0P5E3J3 V9IBT9 A0A0P5GMD5 A0A226EKU6 A0A0P6HUV1 A0A0P6B2C4 A0A1D5NWE6 A0A224YYV0
PDB
2XWE
E-value=3.1821e-25,
Score=280
Ontologies
GO
PANTHER
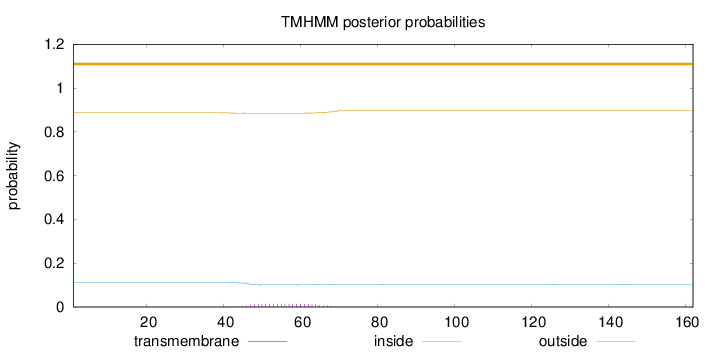
Topology
Length:
162
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31701
Exp number, first 60 AAs:
0.215
Total prob of N-in:
0.11300
outside
1 - 162
Population Genetic Test Statistics
Pi
243.669468
Theta
162.380857
Tajima's D
1.638119
CLR
0.05787
CSRT
0.823558822058897
Interpretation
Uncertain
