Pre Gene Modal
BGIBMGA013129
Annotation
PREDICTED:_glucosylceramidase-like_isoform_X2_[Papilio_xuthus]
Full name
Glucosylceramidase
Location in the cell
PlasmaMembrane Reliability : 2.324
Sequence
CDS
ATGAAAATTTTAATTTGCATTTCTTTGTTAATATTCTACGTATCTTTGAAAGGAGCTCTTTCTGATACCGTATGCCAGGCGAGAGGATCGGATGCGATTGTTTGTGTTTGTAACGACACGTATTGCGATACTGTGACGAGGGAACCTCCGATTATAGGCGCATATAACACCTACACCACATCCGAGAGCGGCTTGCGATTCAGCAAAACTGGAGGAACATTAAACCCGTATAATAATTTAACGCAATGTGGGACGACATTAGTAATAGAACCAAGAGTGAGACATCAAATGATCGAGGGCTTCGGTGGGGCCGTCACTGACGCTGCCGGTATTCACTGGAAGAGCTTGTCGCAAAACTTGCAGAATCATCTTATCGAGTCATACTTCAGCCGCAACGGTCTTGAGTATAACATGATACGCGTTCCGATCGGTGGATCGGATTTCTCAACTCATCCGTACGCATACAACGAATTCCCGGAAAACGATGCAACGCTAAGTAACTTCACACTAGCTAAAGAAGACTACCAGTATAAGATTCCATTGATAAAATCGGCAATGAGTGCCACAAAGAGACAGATCCACATAGTCGCTACGACCTGGTCTCCGCCACCTTGGATGAAAACCAACAACCGGTATTCGGGTATCAGCAGGCTCAAACCTGAATATTTCCAGACCTACGCTGACTACCATCTGAAATTTATACAGAAGTACGCTGATGAAGGAATTCCGGTTTGGGCGATCACAACCACGAACGAACCCTTGTCAGGGATGCTTGGCTTGGACTCTTTTAATACTCTAGGATGGACACAAGATCAAATGGCAAAATGGATAATAAATAATTTAGGACCGACAATTAGGAATTCGACGCACAAAAATACAAAAATACTAGCTGTTGATGATCAGAGAATATCAATACCTTACTGGTTTAACTATATGGTCGCAAAGAACCCGAAGACCTTGGATTATATAGACGGTATAGCCGTGCATTACTACACAGATTTCATCACCCCTGCCTATATATTGACAGAGGTCTCCAAAGCGTTTCCTGATAAGTTTATTTTGGCTACCGAAGCATGTGAAGGAAGTTTCCCGTGGCAGAAGAATAAAGTGGAGCTCGGTTCCTGGACGAGGGCTGCAAACTATATAAACGACATTATAACAGATCTAAATCATAATGTGGTCGGCTGGATAGATTGGAATTTGTGTTTAGACGTCAATGGCGGGCCGAATTGGGCCAAGAACTTCGTTGATTCCCCGATCATAGTTTTCCCCGAAAATGGGGAGTTTGTGAAGCAGCCGATGTTCTATGCGATCGGTCACTTCTCAAAGTTCATACCAAGAGGTTCAGTGCGGATCAAGGTTCATGAAAGAAAAAGAATCTTCTCGAAATCAATAGAAAATGTAGCTTTCTTAACGCCATACGGTACCATTGTTGCTGTGCTTTATAACAGAGGCAACAGAGCTTCCGTGAAAATAAAAATACGAACGAAAGAGATTGTATTAGAATTATTAGCAAAATCGATCAGTACAGTGGAATTTCCCTATAAAACATAG
Protein
MKILICISLLIFYVSLKGALSDTVCQARGSDAIVCVCNDTYCDTVTREPPIIGAYNTYTTSESGLRFSKTGGTLNPYNNLTQCGTTLVIEPRVRHQMIEGFGGAVTDAAGIHWKSLSQNLQNHLIESYFSRNGLEYNMIRVPIGGSDFSTHPYAYNEFPENDATLSNFTLAKEDYQYKIPLIKSAMSATKRQIHIVATTWSPPPWMKTNNRYSGISRLKPEYFQTYADYHLKFIQKYADEGIPVWAITTTNEPLSGMLGLDSFNTLGWTQDQMAKWIINNLGPTIRNSTHKNTKILAVDDQRISIPYWFNYMVAKNPKTLDYIDGIAVHYYTDFITPAYILTEVSKAFPDKFILATEACEGSFPWQKNKVELGSWTRAANYINDIITDLNHNVVGWIDWNLCLDVNGGPNWAKNFVDSPIIVFPENGEFVKQPMFYAIGHFSKFIPRGSVRIKVHERKRIFSKSIENVAFLTPYGTIVAVLYNRGNRASVKIKIRTKEIVLELLAKSISTVEFPYKT
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
H9JUB6
A0A2H1VUS0
A0A194PPY1
A0A437BUD8
A0A1W1EGM7
A0A437B0B5
+ More
A0A194Q614 A0A2A4K975 A0A2H1VG50 A0A212F248 A0A212F254 A0A2H1X380 A0A067QM95 A0A212F293 A0A0C9QXG4 A0A0N1IF15 A0A2H1VST3 A0A1B6FTD6 A0A2J7R6J1 A0A1B6IGC1 A0A1B6K1N7 H9JUB7 A0A194RBR5 A0A154P4F4 D6WRN0 A0A2A4K8P8 A0A1W2W3C7 A0A139WEE1 A0A0L7QMS7 A0A2H1WWW7 L7M0C6 H2YMC3 F7AA01 H2XQ11 A0A088APM4 A0A0P5V115 A0A0P5D4A0 T1JDD3 A0A0P6CTA3 A0A1B6CTI6 A0A182YB56 K7IX35 A0A182PGK5 A0A0P5V259 A0A0P5BE18 A0A0P6HWY3 A0A2A3EMA9 A0A182WSK9 A0A232EVH8 A0A182HLG2 A0A182SVA6 A0A182U728 Q7PXX9 A0A182L9U2 F6VKK2 A0A0P5VAG5 A0A212FNA8 A0A2D4IM94 U3DIF4 A0A210PEG6 U3DTR8 A0A131YKA6 A0A182US45 F7E7C1 A0A182MB91 A0A226EKU6 A0A2K5QV83 A0A250YJK2 A0A2K6SNW8 A0A3S2NLJ3 A0A162QRA1 V4B2P4 A0A2K5QVB6 A0A224YYV0 U3FUA9 A0A2K6ABH7 A0A158NXS0 A0A0N8C3M9 A0A0P5XL26 A0A2C9K3D4 A0A0N8A576 D6WRR5 A0A0P5WP50 A0A1Z5L9E9 K9J1N7 A0A0P5BMC7 A0A1D2MYE0 H3AB74 A0A0P5Z7Y2 H3AB73 A0A139WEN3 A0A034VZ93 A0A3Q7ULG3 A0A3Q7VSS4 L9KHN0 A0A182VZF3 A0A154P6S3 A0A0K8U5E6 A0A0P5X120
A0A194Q614 A0A2A4K975 A0A2H1VG50 A0A212F248 A0A212F254 A0A2H1X380 A0A067QM95 A0A212F293 A0A0C9QXG4 A0A0N1IF15 A0A2H1VST3 A0A1B6FTD6 A0A2J7R6J1 A0A1B6IGC1 A0A1B6K1N7 H9JUB7 A0A194RBR5 A0A154P4F4 D6WRN0 A0A2A4K8P8 A0A1W2W3C7 A0A139WEE1 A0A0L7QMS7 A0A2H1WWW7 L7M0C6 H2YMC3 F7AA01 H2XQ11 A0A088APM4 A0A0P5V115 A0A0P5D4A0 T1JDD3 A0A0P6CTA3 A0A1B6CTI6 A0A182YB56 K7IX35 A0A182PGK5 A0A0P5V259 A0A0P5BE18 A0A0P6HWY3 A0A2A3EMA9 A0A182WSK9 A0A232EVH8 A0A182HLG2 A0A182SVA6 A0A182U728 Q7PXX9 A0A182L9U2 F6VKK2 A0A0P5VAG5 A0A212FNA8 A0A2D4IM94 U3DIF4 A0A210PEG6 U3DTR8 A0A131YKA6 A0A182US45 F7E7C1 A0A182MB91 A0A226EKU6 A0A2K5QV83 A0A250YJK2 A0A2K6SNW8 A0A3S2NLJ3 A0A162QRA1 V4B2P4 A0A2K5QVB6 A0A224YYV0 U3FUA9 A0A2K6ABH7 A0A158NXS0 A0A0N8C3M9 A0A0P5XL26 A0A2C9K3D4 A0A0N8A576 D6WRR5 A0A0P5WP50 A0A1Z5L9E9 K9J1N7 A0A0P5BMC7 A0A1D2MYE0 H3AB74 A0A0P5Z7Y2 H3AB73 A0A139WEN3 A0A034VZ93 A0A3Q7ULG3 A0A3Q7VSS4 L9KHN0 A0A182VZF3 A0A154P6S3 A0A0K8U5E6 A0A0P5X120
EC Number
3.2.1.45
Pubmed
EMBL
BABH01038539
BABH01038540
BABH01038541
ODYU01004571
SOQ44585.1
KQ459602
+ More
KPI93190.1 RSAL01000008 RVE53975.1 LT635665 SGZ49395.1 RSAL01000232 RVE43819.1 KQ459439 KPJ00977.1 NWSH01000035 PCG80448.1 ODYU01002190 SOQ39362.1 AGBW02010783 OWR47794.1 OWR47828.1 ODYU01013051 SOQ59702.1 KK853153 KDR10552.1 OWR47849.1 GBYB01005352 JAG75119.1 KQ461178 KPJ07773.1 ODYU01004232 SOQ43893.1 GECZ01016297 JAS53472.1 NEVH01006756 PNF36452.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 KQ460397 KPJ15278.1 KQ434814 KZC06829.1 KQ971354 EFA07058.2 PCG80449.1 KYB26348.1 KQ414868 KOC59952.1 ODYU01011672 SOQ57565.1 GACK01007712 JAA57322.1 EAAA01001913 GDIP01105548 JAL98166.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 JH432104 GDIP01009481 JAM94234.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 AAZX01006377 AAZX01006727 GDIP01105549 JAL98165.1 GDIP01190080 JAJ33322.1 GDIQ01028607 JAN66130.1 KZ288219 PBC32336.1 NNAY01001997 OXU22351.1 APCN01000897 AAAB01008987 EAA00933.5 GDIP01102691 JAM01024.1 AGBW02006772 OWR55189.1 IACK01117848 LAA85338.1 GAMR01001910 JAB32022.1 NEDP02076748 OWF34883.1 GAMT01003193 GAMS01007908 GAMQ01006239 JAB08668.1 JAB15228.1 JAB35612.1 GEDV01009569 JAP78988.1 AXCM01012486 LNIX01000003 OXA58323.1 GFFW01001008 JAV43780.1 RVE53974.1 KZS19887.1 KB200701 ESP00722.1 GFPF01008008 MAA19154.1 GAMP01004625 JAB48130.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GDIQ01118027 JAL33699.1 GDIP01070494 JAM33221.1 GDIP01181329 JAJ42073.1 EFA05962.2 GDIP01083544 JAM20171.1 GFJQ02003035 JAW03935.1 GABZ01005456 JAA48069.1 GDIP01182978 JAJ40424.1 LJIJ01000420 ODM97675.1 AFYH01203350 AFYH01203351 AFYH01203352 GDIP01060297 JAM43418.1 KYB26349.1 GAKP01011233 JAC47719.1 KB320819 ELW62425.1 KZC06830.1 GDHF01030553 JAI21761.1 GDIP01079198 JAM24517.1
KPI93190.1 RSAL01000008 RVE53975.1 LT635665 SGZ49395.1 RSAL01000232 RVE43819.1 KQ459439 KPJ00977.1 NWSH01000035 PCG80448.1 ODYU01002190 SOQ39362.1 AGBW02010783 OWR47794.1 OWR47828.1 ODYU01013051 SOQ59702.1 KK853153 KDR10552.1 OWR47849.1 GBYB01005352 JAG75119.1 KQ461178 KPJ07773.1 ODYU01004232 SOQ43893.1 GECZ01016297 JAS53472.1 NEVH01006756 PNF36452.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 KQ460397 KPJ15278.1 KQ434814 KZC06829.1 KQ971354 EFA07058.2 PCG80449.1 KYB26348.1 KQ414868 KOC59952.1 ODYU01011672 SOQ57565.1 GACK01007712 JAA57322.1 EAAA01001913 GDIP01105548 JAL98166.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 JH432104 GDIP01009481 JAM94234.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 AAZX01006377 AAZX01006727 GDIP01105549 JAL98165.1 GDIP01190080 JAJ33322.1 GDIQ01028607 JAN66130.1 KZ288219 PBC32336.1 NNAY01001997 OXU22351.1 APCN01000897 AAAB01008987 EAA00933.5 GDIP01102691 JAM01024.1 AGBW02006772 OWR55189.1 IACK01117848 LAA85338.1 GAMR01001910 JAB32022.1 NEDP02076748 OWF34883.1 GAMT01003193 GAMS01007908 GAMQ01006239 JAB08668.1 JAB15228.1 JAB35612.1 GEDV01009569 JAP78988.1 AXCM01012486 LNIX01000003 OXA58323.1 GFFW01001008 JAV43780.1 RVE53974.1 KZS19887.1 KB200701 ESP00722.1 GFPF01008008 MAA19154.1 GAMP01004625 JAB48130.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GDIQ01118027 JAL33699.1 GDIP01070494 JAM33221.1 GDIP01181329 JAJ42073.1 EFA05962.2 GDIP01083544 JAM20171.1 GFJQ02003035 JAW03935.1 GABZ01005456 JAA48069.1 GDIP01182978 JAJ40424.1 LJIJ01000420 ODM97675.1 AFYH01203350 AFYH01203351 AFYH01203352 GDIP01060297 JAM43418.1 KYB26349.1 GAKP01011233 JAC47719.1 KB320819 ELW62425.1 KZC06830.1 GDHF01030553 JAI21761.1 GDIP01079198 JAM24517.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000007151
UP000027135
+ More
UP000053240 UP000235965 UP000076502 UP000007266 UP000053825 UP000007875 UP000008144 UP000005203 UP000076858 UP000076408 UP000002358 UP000075885 UP000242457 UP000076407 UP000215335 UP000075840 UP000075901 UP000075902 UP000007062 UP000075882 UP000002280 UP000242188 UP000075903 UP000008225 UP000075883 UP000198287 UP000233040 UP000233220 UP000030746 UP000233140 UP000005205 UP000076420 UP000094527 UP000008672 UP000286642 UP000011518 UP000075920
UP000053240 UP000235965 UP000076502 UP000007266 UP000053825 UP000007875 UP000008144 UP000005203 UP000076858 UP000076408 UP000002358 UP000075885 UP000242457 UP000076407 UP000215335 UP000075840 UP000075901 UP000075902 UP000007062 UP000075882 UP000002280 UP000242188 UP000075903 UP000008225 UP000075883 UP000198287 UP000233040 UP000233220 UP000030746 UP000233140 UP000005205 UP000076420 UP000094527 UP000008672 UP000286642 UP000011518 UP000075920
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JUB6
A0A2H1VUS0
A0A194PPY1
A0A437BUD8
A0A1W1EGM7
A0A437B0B5
+ More
A0A194Q614 A0A2A4K975 A0A2H1VG50 A0A212F248 A0A212F254 A0A2H1X380 A0A067QM95 A0A212F293 A0A0C9QXG4 A0A0N1IF15 A0A2H1VST3 A0A1B6FTD6 A0A2J7R6J1 A0A1B6IGC1 A0A1B6K1N7 H9JUB7 A0A194RBR5 A0A154P4F4 D6WRN0 A0A2A4K8P8 A0A1W2W3C7 A0A139WEE1 A0A0L7QMS7 A0A2H1WWW7 L7M0C6 H2YMC3 F7AA01 H2XQ11 A0A088APM4 A0A0P5V115 A0A0P5D4A0 T1JDD3 A0A0P6CTA3 A0A1B6CTI6 A0A182YB56 K7IX35 A0A182PGK5 A0A0P5V259 A0A0P5BE18 A0A0P6HWY3 A0A2A3EMA9 A0A182WSK9 A0A232EVH8 A0A182HLG2 A0A182SVA6 A0A182U728 Q7PXX9 A0A182L9U2 F6VKK2 A0A0P5VAG5 A0A212FNA8 A0A2D4IM94 U3DIF4 A0A210PEG6 U3DTR8 A0A131YKA6 A0A182US45 F7E7C1 A0A182MB91 A0A226EKU6 A0A2K5QV83 A0A250YJK2 A0A2K6SNW8 A0A3S2NLJ3 A0A162QRA1 V4B2P4 A0A2K5QVB6 A0A224YYV0 U3FUA9 A0A2K6ABH7 A0A158NXS0 A0A0N8C3M9 A0A0P5XL26 A0A2C9K3D4 A0A0N8A576 D6WRR5 A0A0P5WP50 A0A1Z5L9E9 K9J1N7 A0A0P5BMC7 A0A1D2MYE0 H3AB74 A0A0P5Z7Y2 H3AB73 A0A139WEN3 A0A034VZ93 A0A3Q7ULG3 A0A3Q7VSS4 L9KHN0 A0A182VZF3 A0A154P6S3 A0A0K8U5E6 A0A0P5X120
A0A194Q614 A0A2A4K975 A0A2H1VG50 A0A212F248 A0A212F254 A0A2H1X380 A0A067QM95 A0A212F293 A0A0C9QXG4 A0A0N1IF15 A0A2H1VST3 A0A1B6FTD6 A0A2J7R6J1 A0A1B6IGC1 A0A1B6K1N7 H9JUB7 A0A194RBR5 A0A154P4F4 D6WRN0 A0A2A4K8P8 A0A1W2W3C7 A0A139WEE1 A0A0L7QMS7 A0A2H1WWW7 L7M0C6 H2YMC3 F7AA01 H2XQ11 A0A088APM4 A0A0P5V115 A0A0P5D4A0 T1JDD3 A0A0P6CTA3 A0A1B6CTI6 A0A182YB56 K7IX35 A0A182PGK5 A0A0P5V259 A0A0P5BE18 A0A0P6HWY3 A0A2A3EMA9 A0A182WSK9 A0A232EVH8 A0A182HLG2 A0A182SVA6 A0A182U728 Q7PXX9 A0A182L9U2 F6VKK2 A0A0P5VAG5 A0A212FNA8 A0A2D4IM94 U3DIF4 A0A210PEG6 U3DTR8 A0A131YKA6 A0A182US45 F7E7C1 A0A182MB91 A0A226EKU6 A0A2K5QV83 A0A250YJK2 A0A2K6SNW8 A0A3S2NLJ3 A0A162QRA1 V4B2P4 A0A2K5QVB6 A0A224YYV0 U3FUA9 A0A2K6ABH7 A0A158NXS0 A0A0N8C3M9 A0A0P5XL26 A0A2C9K3D4 A0A0N8A576 D6WRR5 A0A0P5WP50 A0A1Z5L9E9 K9J1N7 A0A0P5BMC7 A0A1D2MYE0 H3AB74 A0A0P5Z7Y2 H3AB73 A0A139WEN3 A0A034VZ93 A0A3Q7ULG3 A0A3Q7VSS4 L9KHN0 A0A182VZF3 A0A154P6S3 A0A0K8U5E6 A0A0P5X120
PDB
2XWE
E-value=1.21836e-109,
Score=1015
Ontologies
PATHWAY
GO
GO:0006665
GO:0004348
GO:0006680
GO:0016787
GO:0016021
GO:0032715
GO:0050295
GO:0032436
GO:0005783
GO:0005615
GO:0046512
GO:0009267
GO:0007040
GO:0032463
GO:0043202
GO:1905037
GO:0005765
GO:0043243
GO:1903061
GO:0023021
GO:0030259
GO:0071356
GO:0005124
GO:0005802
GO:0046527
GO:1904457
GO:0046513
GO:0035307
GO:1904925
GO:0032006
GO:0008203
GO:1901215
GO:0043407
GO:1903052
GO:0006914
GO:0005975
GO:0008270
GO:0005488
GO:0008565
GO:0015031
GO:0009258
GO:0016155
GO:0003824
PANTHER
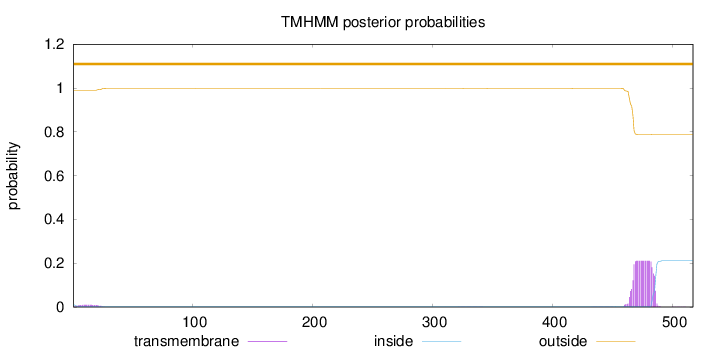
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.998903
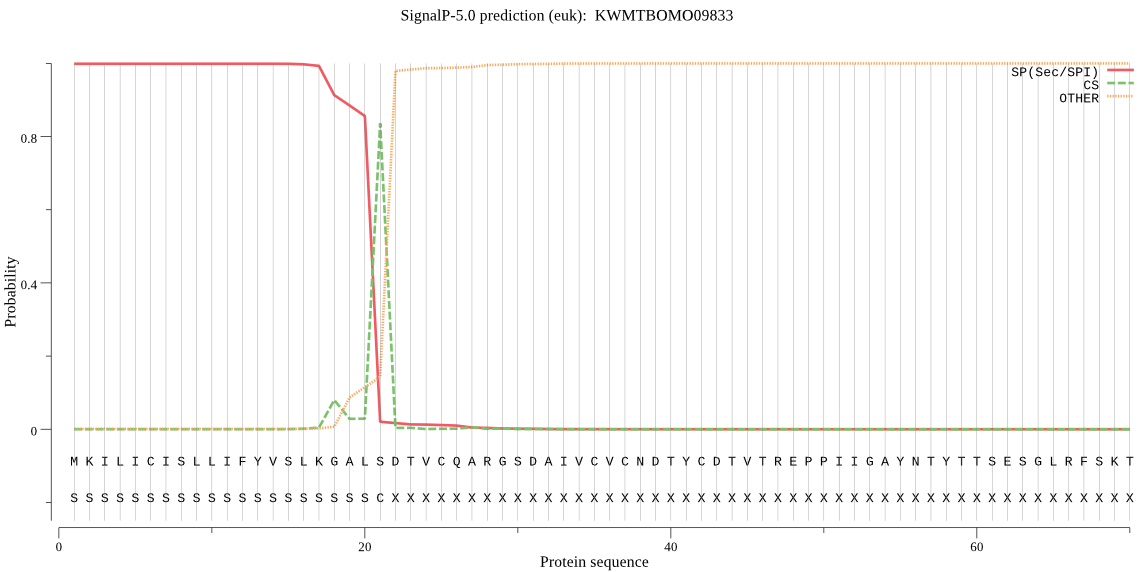
Length:
517
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.28765
Exp number, first 60 AAs:
0.18378
Total prob of N-in:
0.01012
outside
1 - 517
Population Genetic Test Statistics
Pi
182.318393
Theta
166.879161
Tajima's D
0.37754
CLR
0.581239
CSRT
0.474176291185441
Interpretation
Uncertain
