Gene
KWMTBOMO09832 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013129
Annotation
PREDICTED:_glucosylceramidase-like_isoform_X3_[Bombyx_mori]
Full name
Glucosylceramidase
Location in the cell
Cytoplasmic Reliability : 1.094 PlasmaMembrane Reliability : 1.317
Sequence
CDS
ATGATCAAACGTAACTTATTAGTTGTGGGATTTGAGATCTCACTGTTGTTTAGCTTTGTTGTTGGATATTATAGGGATAAAACATGCGCCGAGCTGCAAGTAAACAATAGCTCCGTGATATGCGTCTGCAATGCAACGTACTGTGACACTGTGACTAGAGAGGAGCCGCACCCTAGAAGATTTATGGTGTATACTTCCTCTAAGGCAGGACTGCGGTTCAAGAAGTTCACATCTATCTTGGAGGTCTTCGATTTGTTAACAGAATGTTCTACAACCTTAGAACTCGAACCCAGAAAGAGGTATCAGACGATCCATGGCTTCGGAGGCGCCATCACCGATTCAGCGGCCATCAATTGGAGCAACTTAAAAGATGAAGAATTAAAGCAGAATTTAATAGAATCCTACTGCGGTGACAGCGGAATACAATACAACATGCTCCGCGTGCCGATCGGTGGAACCGACTTCTCACCTCGCTTCTACACATACCAGGACTACCCCGAGAGAGACAAGGACTTAAGCAACTTCACACTTGCTCCCGAGGACTTGAACTATAAGCTGCCGATGATAAAAGCCTGCATGAAAGCGTCAAATGCCTCAATAGATATCCTAGGATCGTTATGGTATCCACCTGCGTGGATGAAGAGTACGAAAAAATCGAACGGAATTTCTTTTCTGGAGGAGGAATATTATGACGCTCTCGCCCTGTATCATCTCAAATTTATCAAAGAGTATTACAAACAGGGCGTTCCGATTTGGGGCATAACTACAACGAATGAACCTTTGAGCGGGATTACAAAACATGCTAATGATAATGTACTCGGCTGGTCGGTTTACGGATTGGCAAATTGGATTGTCAATTATTTGGCACCAAAAATAAAACGGTACGATCCAAGCATCAAGATACTAGGAGTTGACGACCAGAGATTTACTGTTCCACTATGGTTCAATGCGGCATTAAAACATGTACCGGAACTAGGGAGAGCTATTGACGGGATCGGACTACATTACTATGAGAACGAACTCGTCCCGCCCGAGATAGTCGACCGATGTCTTAAAAATTATCCTAATCTGTTCGTTTTGTCTTCAGAAGCATCTATACATTCACAAACAAATAATAAAGTAGATTTAGGATCGTGGGATCGAGCTGAAAAATACATTAAGAATATAATTGAGGACCTAAATCACAACTACACAGCCTGGATCGATTGGAACGTCTGTCTTGACAAAATGGGGGGGCCTGTTTTGTCCAAAACCTATCTGGACTCGCCCGTGATTGTGGACCCTGATGCAGGGGTATTCTACAAACAGCCGTTGTTCTACGCTATGGGTCACTTCTCAAAGTTCATCCCTAAAGGTTCTAAAAGGATCCAGTCCATGGAGAAGTACAATTGTGGGGATCCCATAAAGCATGTAGCGTTTTCGACTCCCATGAATACAATAGTTGTCGTTGTTTTTAATGATAAGAATAAAGAGAAAGTGGTACGTCTGAAGCTGGGATACAGCCAAGCGAAGGTCCTGTTAGAGAAGAATTCGATTGCGACAGTCGAATTTCATAATGACGTAATCGACGAATCATGTGACTGTGAGAAGTAG
Protein
MIKRNLLVVGFEISLLFSFVVGYYRDKTCAELQVNNSSVICVCNATYCDTVTREEPHPRRFMVYTSSKAGLRFKKFTSILEVFDLLTECSTTLELEPRKRYQTIHGFGGAITDSAAINWSNLKDEELKQNLIESYCGDSGIQYNMLRVPIGGTDFSPRFYTYQDYPERDKDLSNFTLAPEDLNYKLPMIKACMKASNASIDILGSLWYPPAWMKSTKKSNGISFLEEEYYDALALYHLKFIKEYYKQGVPIWGITTTNEPLSGITKHANDNVLGWSVYGLANWIVNYLAPKIKRYDPSIKILGVDDQRFTVPLWFNAALKHVPELGRAIDGIGLHYYENELVPPEIVDRCLKNYPNLFVLSSEASIHSQTNNKVDLGSWDRAEKYIKNIIEDLNHNYTAWIDWNVCLDKMGGPVLSKTYLDSPVIVDPDAGVFYKQPLFYAMGHFSKFIPKGSKRIQSMEKYNCGDPIKHVAFSTPMNTIVVVVFNDKNKEKVVRLKLGYSQAKVLLEKNSIATVEFHNDVIDESCDCEK
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
H9JUB6
H9JUB7
A0A2A4K975
A0A2H1VUS0
A0A1W1EGM7
A0A194PPY1
+ More
A0A212F254 A0A437BUD8 A0A2H1X380 A0A2H1VST3 A0A212F248 A0A067QM95 A0A437B0B5 A0A212F293 A0A194Q614 A0A139WEE1 A0A0N1IF15 A0A0C9QXG4 A0A2H1VG50 D6WRN0 A0A2J7R6J1 A0A139WEN3 A0A131YKA6 A0A2H1WWW7 A0A224YYV0 A0A131Z3G7 A0A224YXV5 L7M0C6 L7M0B5 A0A1E1X9A0 A0A1B6FTD6 A0A1B6IGC1 D6WRR5 A0A1B6K1N7 A0A151WMN1 A0A088APM4 A0A154P4F4 A0A1W4WRP8 A0A1W2W3C7 A0A2A3EMA9 E0VQX9 A0A158NXS0 F4X220 H2YMC3 A0A1B6CTI6 F7AA01 A0A131YJ49 D6WRR4 A0A195ECY0 A0A232EVH8 A0A1W2WHS5 A0A224YX30 A0A1Y1L8U4 A0A1L8FDF0 H2YNM8 H2XJY8 J3JXY3 H2XQ11 K7IX35 A0A1V9X5I8 A0A195B051 A0A0L7QMS7 A0A0P5V115 A0A1Z5LER5 A0A088APM5 V4B2P4 A0A131ZAG4 A0A1Z5L9E9 A0A016WL53 A0A226EKU6 C3YQ03 A0A0P5D4A0 A0A0P6HWY3 A0A0P6CTA3 A0A016WKE4 A0A0P5V259 A0A0P5XL26 A0A154P6S3 A0A0P5BE18 A0A0P5MXS5 A0A1S3HF23 A0A368H7B4 A0A3Q4GE16 A0A2A3EKU2 A0A0P6E3A7 A0A1S0U9S6 E7EZM1 A0A210PEG6 A0A162QRA1 A0A0P5VAG5 A0A0N8C3M9 A0A195CKU4 V9IJV6 A0A3P6TRN0 A0A423SIH3 A0A1D2MYE0 A0A0N8A576 A0A026WL55 A0A0P5BMC7
A0A212F254 A0A437BUD8 A0A2H1X380 A0A2H1VST3 A0A212F248 A0A067QM95 A0A437B0B5 A0A212F293 A0A194Q614 A0A139WEE1 A0A0N1IF15 A0A0C9QXG4 A0A2H1VG50 D6WRN0 A0A2J7R6J1 A0A139WEN3 A0A131YKA6 A0A2H1WWW7 A0A224YYV0 A0A131Z3G7 A0A224YXV5 L7M0C6 L7M0B5 A0A1E1X9A0 A0A1B6FTD6 A0A1B6IGC1 D6WRR5 A0A1B6K1N7 A0A151WMN1 A0A088APM4 A0A154P4F4 A0A1W4WRP8 A0A1W2W3C7 A0A2A3EMA9 E0VQX9 A0A158NXS0 F4X220 H2YMC3 A0A1B6CTI6 F7AA01 A0A131YJ49 D6WRR4 A0A195ECY0 A0A232EVH8 A0A1W2WHS5 A0A224YX30 A0A1Y1L8U4 A0A1L8FDF0 H2YNM8 H2XJY8 J3JXY3 H2XQ11 K7IX35 A0A1V9X5I8 A0A195B051 A0A0L7QMS7 A0A0P5V115 A0A1Z5LER5 A0A088APM5 V4B2P4 A0A131ZAG4 A0A1Z5L9E9 A0A016WL53 A0A226EKU6 C3YQ03 A0A0P5D4A0 A0A0P6HWY3 A0A0P6CTA3 A0A016WKE4 A0A0P5V259 A0A0P5XL26 A0A154P6S3 A0A0P5BE18 A0A0P5MXS5 A0A1S3HF23 A0A368H7B4 A0A3Q4GE16 A0A2A3EKU2 A0A0P6E3A7 A0A1S0U9S6 E7EZM1 A0A210PEG6 A0A162QRA1 A0A0P5VAG5 A0A0N8C3M9 A0A195CKU4 V9IJV6 A0A3P6TRN0 A0A423SIH3 A0A1D2MYE0 A0A0N8A576 A0A026WL55 A0A0P5BMC7
EC Number
3.2.1.45
Pubmed
EMBL
BABH01038539
BABH01038540
BABH01038541
BABH01038531
BABH01038532
BABH01038533
+ More
BABH01038534 NWSH01000035 PCG80448.1 ODYU01004571 SOQ44585.1 LT635665 SGZ49395.1 KQ459602 KPI93190.1 AGBW02010783 OWR47828.1 RSAL01000008 RVE53975.1 ODYU01013051 SOQ59702.1 ODYU01004232 SOQ43893.1 OWR47794.1 KK853153 KDR10552.1 RSAL01000232 RVE43819.1 OWR47849.1 KQ459439 KPJ00977.1 KQ971354 KYB26348.1 KQ461178 KPJ07773.1 GBYB01005352 JAG75119.1 ODYU01002190 SOQ39362.1 EFA07058.2 NEVH01006756 PNF36452.1 KYB26349.1 GEDV01009569 JAP78988.1 ODYU01011672 SOQ57565.1 GFPF01008008 MAA19154.1 GEDV01003886 JAP84671.1 GFPF01008007 MAA19153.1 GACK01007712 JAA57322.1 GACK01007727 JAA57307.1 GFAC01003537 JAT95651.1 GECZ01016297 JAS53472.1 GECU01021735 JAS85971.1 EFA05962.2 GECU01002337 JAT05370.1 KQ982934 KYQ49129.1 KQ434814 KZC06829.1 KZ288219 PBC32336.1 DS235442 EEB15785.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GL888561 EGI59502.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 EAAA01001913 GEDV01009570 JAP78987.1 EFA05963.2 KQ979074 KYN23063.1 NNAY01001997 OXU22351.1 GFPF01008005 MAA19151.1 GEZM01063170 JAV69258.1 CM004480 OCT69622.1 EAAA01002615 BT128105 AEE63066.1 AAZX01006377 AAZX01006727 MNPL01023560 OQR68676.1 KQ976692 KYM77670.1 KQ414868 KOC59952.1 GDIP01105548 JAL98166.1 GFJQ02001071 JAW05899.1 KB200701 ESP00722.1 GEDV01001091 JAP87466.1 GFJQ02003035 JAW03935.1 JARK01000241 EYC39763.1 LNIX01000003 OXA58323.1 GG666539 EEN57641.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01028607 JAN66130.1 GDIP01009481 JAM94234.1 KE124952 EPB74230.1 EYC39762.1 GDIP01105549 JAL98165.1 GDIP01070494 JAM33221.1 KZC06830.1 GDIP01190080 JAJ33322.1 GDIQ01149727 JAL01999.1 JOJR01000007 RCN52482.1 PBC32337.1 GDIQ01068405 JAN26332.1 JH712071 EFO26573.2 BX322530 NEDP02076748 OWF34883.1 KZS19887.1 GDIP01102691 JAM01024.1 GDIQ01118027 JAL33699.1 KQ977622 KYN01328.1 JR049360 AEY60937.1 UYRX01000558 VDK83835.1 QCYY01003347 ROT63991.1 LJIJ01000420 ODM97675.1 GDIP01181329 JAJ42073.1 KK107159 QOIP01000001 EZA56663.1 RLU27086.1 GDIP01182978 JAJ40424.1
BABH01038534 NWSH01000035 PCG80448.1 ODYU01004571 SOQ44585.1 LT635665 SGZ49395.1 KQ459602 KPI93190.1 AGBW02010783 OWR47828.1 RSAL01000008 RVE53975.1 ODYU01013051 SOQ59702.1 ODYU01004232 SOQ43893.1 OWR47794.1 KK853153 KDR10552.1 RSAL01000232 RVE43819.1 OWR47849.1 KQ459439 KPJ00977.1 KQ971354 KYB26348.1 KQ461178 KPJ07773.1 GBYB01005352 JAG75119.1 ODYU01002190 SOQ39362.1 EFA07058.2 NEVH01006756 PNF36452.1 KYB26349.1 GEDV01009569 JAP78988.1 ODYU01011672 SOQ57565.1 GFPF01008008 MAA19154.1 GEDV01003886 JAP84671.1 GFPF01008007 MAA19153.1 GACK01007712 JAA57322.1 GACK01007727 JAA57307.1 GFAC01003537 JAT95651.1 GECZ01016297 JAS53472.1 GECU01021735 JAS85971.1 EFA05962.2 GECU01002337 JAT05370.1 KQ982934 KYQ49129.1 KQ434814 KZC06829.1 KZ288219 PBC32336.1 DS235442 EEB15785.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GL888561 EGI59502.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 EAAA01001913 GEDV01009570 JAP78987.1 EFA05963.2 KQ979074 KYN23063.1 NNAY01001997 OXU22351.1 GFPF01008005 MAA19151.1 GEZM01063170 JAV69258.1 CM004480 OCT69622.1 EAAA01002615 BT128105 AEE63066.1 AAZX01006377 AAZX01006727 MNPL01023560 OQR68676.1 KQ976692 KYM77670.1 KQ414868 KOC59952.1 GDIP01105548 JAL98166.1 GFJQ02001071 JAW05899.1 KB200701 ESP00722.1 GEDV01001091 JAP87466.1 GFJQ02003035 JAW03935.1 JARK01000241 EYC39763.1 LNIX01000003 OXA58323.1 GG666539 EEN57641.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01028607 JAN66130.1 GDIP01009481 JAM94234.1 KE124952 EPB74230.1 EYC39762.1 GDIP01105549 JAL98165.1 GDIP01070494 JAM33221.1 KZC06830.1 GDIP01190080 JAJ33322.1 GDIQ01149727 JAL01999.1 JOJR01000007 RCN52482.1 PBC32337.1 GDIQ01068405 JAN26332.1 JH712071 EFO26573.2 BX322530 NEDP02076748 OWF34883.1 KZS19887.1 GDIP01102691 JAM01024.1 GDIQ01118027 JAL33699.1 KQ977622 KYN01328.1 JR049360 AEY60937.1 UYRX01000558 VDK83835.1 QCYY01003347 ROT63991.1 LJIJ01000420 ODM97675.1 GDIP01181329 JAJ42073.1 KK107159 QOIP01000001 EZA56663.1 RLU27086.1 GDIP01182978 JAJ40424.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000027135
+ More
UP000007266 UP000053240 UP000235965 UP000075809 UP000005203 UP000076502 UP000192223 UP000242457 UP000009046 UP000005205 UP000007755 UP000007875 UP000008144 UP000078492 UP000215335 UP000186698 UP000002358 UP000192247 UP000078540 UP000053825 UP000030746 UP000024635 UP000198287 UP000001554 UP000076858 UP000085678 UP000252519 UP000261580 UP000000437 UP000242188 UP000078542 UP000277928 UP000283509 UP000094527 UP000053097 UP000279307
UP000007266 UP000053240 UP000235965 UP000075809 UP000005203 UP000076502 UP000192223 UP000242457 UP000009046 UP000005205 UP000007755 UP000007875 UP000008144 UP000078492 UP000215335 UP000186698 UP000002358 UP000192247 UP000078540 UP000053825 UP000030746 UP000024635 UP000198287 UP000001554 UP000076858 UP000085678 UP000252519 UP000261580 UP000000437 UP000242188 UP000078542 UP000277928 UP000283509 UP000094527 UP000053097 UP000279307
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JUB6
H9JUB7
A0A2A4K975
A0A2H1VUS0
A0A1W1EGM7
A0A194PPY1
+ More
A0A212F254 A0A437BUD8 A0A2H1X380 A0A2H1VST3 A0A212F248 A0A067QM95 A0A437B0B5 A0A212F293 A0A194Q614 A0A139WEE1 A0A0N1IF15 A0A0C9QXG4 A0A2H1VG50 D6WRN0 A0A2J7R6J1 A0A139WEN3 A0A131YKA6 A0A2H1WWW7 A0A224YYV0 A0A131Z3G7 A0A224YXV5 L7M0C6 L7M0B5 A0A1E1X9A0 A0A1B6FTD6 A0A1B6IGC1 D6WRR5 A0A1B6K1N7 A0A151WMN1 A0A088APM4 A0A154P4F4 A0A1W4WRP8 A0A1W2W3C7 A0A2A3EMA9 E0VQX9 A0A158NXS0 F4X220 H2YMC3 A0A1B6CTI6 F7AA01 A0A131YJ49 D6WRR4 A0A195ECY0 A0A232EVH8 A0A1W2WHS5 A0A224YX30 A0A1Y1L8U4 A0A1L8FDF0 H2YNM8 H2XJY8 J3JXY3 H2XQ11 K7IX35 A0A1V9X5I8 A0A195B051 A0A0L7QMS7 A0A0P5V115 A0A1Z5LER5 A0A088APM5 V4B2P4 A0A131ZAG4 A0A1Z5L9E9 A0A016WL53 A0A226EKU6 C3YQ03 A0A0P5D4A0 A0A0P6HWY3 A0A0P6CTA3 A0A016WKE4 A0A0P5V259 A0A0P5XL26 A0A154P6S3 A0A0P5BE18 A0A0P5MXS5 A0A1S3HF23 A0A368H7B4 A0A3Q4GE16 A0A2A3EKU2 A0A0P6E3A7 A0A1S0U9S6 E7EZM1 A0A210PEG6 A0A162QRA1 A0A0P5VAG5 A0A0N8C3M9 A0A195CKU4 V9IJV6 A0A3P6TRN0 A0A423SIH3 A0A1D2MYE0 A0A0N8A576 A0A026WL55 A0A0P5BMC7
A0A212F254 A0A437BUD8 A0A2H1X380 A0A2H1VST3 A0A212F248 A0A067QM95 A0A437B0B5 A0A212F293 A0A194Q614 A0A139WEE1 A0A0N1IF15 A0A0C9QXG4 A0A2H1VG50 D6WRN0 A0A2J7R6J1 A0A139WEN3 A0A131YKA6 A0A2H1WWW7 A0A224YYV0 A0A131Z3G7 A0A224YXV5 L7M0C6 L7M0B5 A0A1E1X9A0 A0A1B6FTD6 A0A1B6IGC1 D6WRR5 A0A1B6K1N7 A0A151WMN1 A0A088APM4 A0A154P4F4 A0A1W4WRP8 A0A1W2W3C7 A0A2A3EMA9 E0VQX9 A0A158NXS0 F4X220 H2YMC3 A0A1B6CTI6 F7AA01 A0A131YJ49 D6WRR4 A0A195ECY0 A0A232EVH8 A0A1W2WHS5 A0A224YX30 A0A1Y1L8U4 A0A1L8FDF0 H2YNM8 H2XJY8 J3JXY3 H2XQ11 K7IX35 A0A1V9X5I8 A0A195B051 A0A0L7QMS7 A0A0P5V115 A0A1Z5LER5 A0A088APM5 V4B2P4 A0A131ZAG4 A0A1Z5L9E9 A0A016WL53 A0A226EKU6 C3YQ03 A0A0P5D4A0 A0A0P6HWY3 A0A0P6CTA3 A0A016WKE4 A0A0P5V259 A0A0P5XL26 A0A154P6S3 A0A0P5BE18 A0A0P5MXS5 A0A1S3HF23 A0A368H7B4 A0A3Q4GE16 A0A2A3EKU2 A0A0P6E3A7 A0A1S0U9S6 E7EZM1 A0A210PEG6 A0A162QRA1 A0A0P5VAG5 A0A0N8C3M9 A0A195CKU4 V9IJV6 A0A3P6TRN0 A0A423SIH3 A0A1D2MYE0 A0A0N8A576 A0A026WL55 A0A0P5BMC7
PDB
2XWE
E-value=3.42689e-76,
Score=726
Ontologies
PATHWAY
GO
PANTHER
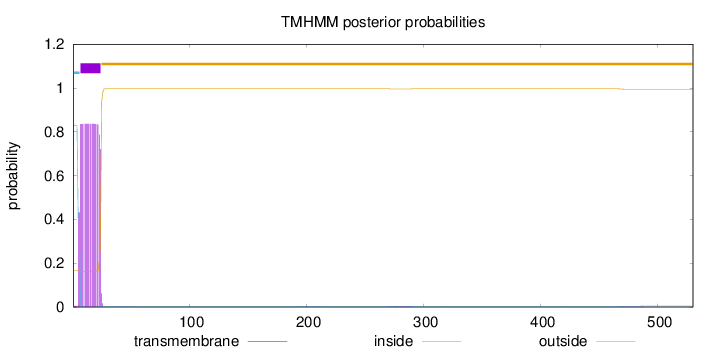
Topology
Length:
530
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.94075
Exp number, first 60 AAs:
15.83974
Total prob of N-in:
0.83334
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 530
Population Genetic Test Statistics
Pi
245.274042
Theta
174.911025
Tajima's D
0.282785
CLR
150.305411
CSRT
0.451827408629568
Interpretation
Uncertain
